Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
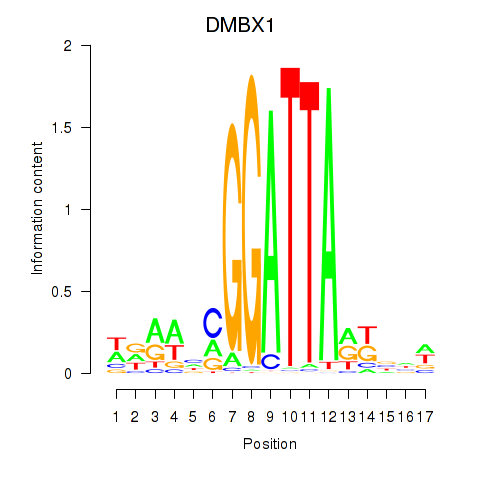
Results for DMBX1
Z-value: 0.32
Transcription factors associated with DMBX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DMBX1
|
ENSG00000197587.6 | diencephalon/mesencephalon homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DMBX1 | hg19_v2_chr1_+_46972668_46972669 | 0.41 | 6.9e-02 | Click! |
Activity profile of DMBX1 motif
Sorted Z-values of DMBX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_223725723 | 0.58 |
ENST00000535678.1
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr2_-_175711133 | 0.38 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr17_-_19651654 | 0.37 |
ENST00000395555.3
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr17_-_19651598 | 0.36 |
ENST00000570414.1
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr8_-_23282797 | 0.35 |
ENST00000524144.1
|
LOXL2
|
lysyl oxidase-like 2 |
| chr17_-_19651668 | 0.34 |
ENST00000494157.2
ENST00000225740.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr8_-_23282820 | 0.34 |
ENST00000520871.1
|
LOXL2
|
lysyl oxidase-like 2 |
| chr2_-_220252068 | 0.33 |
ENST00000430206.1
ENST00000429013.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr3_+_111393501 | 0.31 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr4_+_169552748 | 0.31 |
ENST00000504519.1
ENST00000512127.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr3_-_149293990 | 0.29 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr17_+_16945820 | 0.28 |
ENST00000577514.1
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr5_+_71403280 | 0.27 |
ENST00000511641.2
|
MAP1B
|
microtubule-associated protein 1B |
| chr3_-_137851220 | 0.26 |
ENST00000236709.3
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr15_+_63414760 | 0.25 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr3_-_11685345 | 0.25 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr1_+_120839005 | 0.24 |
ENST00000369390.3
ENST00000452190.1 |
FAM72B
|
family with sequence similarity 72, member B |
| chr1_-_143913143 | 0.24 |
ENST00000400889.1
|
FAM72D
|
family with sequence similarity 72, member D |
| chr1_+_206138457 | 0.23 |
ENST00000367128.3
ENST00000431655.2 |
FAM72A
|
family with sequence similarity 72, member A |
| chr16_-_31076332 | 0.23 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr6_-_33297013 | 0.22 |
ENST00000453407.1
|
DAXX
|
death-domain associated protein |
| chr15_-_60771280 | 0.22 |
ENST00000560072.1
ENST00000560406.1 ENST00000560520.1 ENST00000261520.4 ENST00000439632.1 |
NARG2
|
NMDA receptor regulated 2 |
| chr2_-_27486951 | 0.21 |
ENST00000432351.1
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr1_-_47655686 | 0.21 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr2_-_220252530 | 0.19 |
ENST00000521459.1
|
DNPEP
|
aspartyl aminopeptidase |
| chr3_+_99986036 | 0.19 |
ENST00000471098.1
|
TBC1D23
|
TBC1 domain family, member 23 |
| chr12_-_76461249 | 0.18 |
ENST00000551524.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr3_-_49058479 | 0.18 |
ENST00000440857.1
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr15_-_85201779 | 0.17 |
ENST00000360476.3
ENST00000394588.3 |
NMB
|
neuromedin B |
| chr15_+_63413990 | 0.17 |
ENST00000261893.4
|
LACTB
|
lactamase, beta |
| chr14_+_50234309 | 0.16 |
ENST00000298307.5
|
KLHDC2
|
kelch domain containing 2 |
| chr2_+_97454321 | 0.16 |
ENST00000540067.1
|
CNNM4
|
cyclin M4 |
| chr10_-_101491828 | 0.16 |
ENST00000370483.5
ENST00000016171.5 |
COX15
|
cytochrome c oxidase assembly homolog 15 (yeast) |
| chr16_-_31076273 | 0.15 |
ENST00000426488.2
|
ZNF668
|
zinc finger protein 668 |
| chrX_-_64754611 | 0.15 |
ENST00000374807.5
ENST00000374811.3 ENST00000374804.5 ENST00000312391.8 |
LAS1L
|
LAS1-like (S. cerevisiae) |
| chr5_-_54988448 | 0.15 |
ENST00000503817.1
ENST00000512595.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr3_-_19975665 | 0.14 |
ENST00000295824.9
ENST00000389256.4 |
EFHB
|
EF-hand domain family, member B |
| chr22_+_31002990 | 0.14 |
ENST00000423350.1
|
TCN2
|
transcobalamin II |
| chr14_-_23395623 | 0.14 |
ENST00000556043.1
|
PRMT5
|
protein arginine methyltransferase 5 |
| chr2_+_37571845 | 0.14 |
ENST00000537448.1
|
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr1_+_81001398 | 0.13 |
ENST00000418041.1
ENST00000443104.1 |
RP5-887A10.1
|
RP5-887A10.1 |
| chr3_+_111393659 | 0.13 |
ENST00000477665.1
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr22_-_43411106 | 0.13 |
ENST00000453643.1
ENST00000263246.3 ENST00000337959.4 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr2_+_73461364 | 0.12 |
ENST00000540468.1
ENST00000539919.1 ENST00000258091.5 |
CCT7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr10_-_99030395 | 0.12 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr8_-_110656995 | 0.12 |
ENST00000276646.9
ENST00000533065.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr2_-_70780572 | 0.11 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chr12_-_49581152 | 0.11 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr3_+_138340067 | 0.11 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr2_+_73461410 | 0.11 |
ENST00000399032.2
ENST00000398422.2 ENST00000537131.1 ENST00000538797.1 |
CCT7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr12_-_118628315 | 0.10 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr1_-_47779762 | 0.10 |
ENST00000371877.3
ENST00000360380.3 ENST00000337817.5 ENST00000447475.2 |
STIL
|
SCL/TAL1 interrupting locus |
| chr6_-_26250835 | 0.10 |
ENST00000446824.2
|
HIST1H3F
|
histone cluster 1, H3f |
| chr2_-_14541060 | 0.10 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr10_-_35104185 | 0.09 |
ENST00000374789.3
ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3
|
par-3 family cell polarity regulator |
| chr1_+_180875711 | 0.09 |
ENST00000434447.1
|
RP11-46A10.2
|
RP11-46A10.2 |
| chr4_-_170679024 | 0.08 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
| chr1_-_16563641 | 0.08 |
ENST00000375599.3
|
RSG1
|
REM2 and RAB-like small GTPase 1 |
| chr9_+_74920408 | 0.08 |
ENST00000451152.1
|
RP11-63P12.6
|
RP11-63P12.6 |
| chr6_+_10585979 | 0.08 |
ENST00000265012.4
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr4_+_41937131 | 0.07 |
ENST00000504986.1
ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33
|
transmembrane protein 33 |
| chr10_+_74451883 | 0.07 |
ENST00000373053.3
ENST00000357157.6 |
MCU
|
mitochondrial calcium uniporter |
| chr9_+_74920335 | 0.07 |
ENST00000451596.2
ENST00000436054.1 |
RP11-63P12.6
|
RP11-63P12.6 |
| chr17_+_60536002 | 0.07 |
ENST00000582809.1
|
TLK2
|
tousled-like kinase 2 |
| chr19_+_17448348 | 0.06 |
ENST00000324894.8
ENST00000358792.7 ENST00000600625.1 |
GTPBP3
|
GTP binding protein 3 (mitochondrial) |
| chr19_-_17186229 | 0.06 |
ENST00000253669.5
ENST00000448593.2 |
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr4_+_108815402 | 0.06 |
ENST00000503385.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr17_-_12921270 | 0.05 |
ENST00000578071.1
ENST00000426905.3 ENST00000395962.2 ENST00000583371.1 ENST00000338034.4 |
ELAC2
|
elaC ribonuclease Z 2 |
| chr9_+_107266455 | 0.05 |
ENST00000334726.2
|
OR13F1
|
olfactory receptor, family 13, subfamily F, member 1 |
| chr19_-_17185848 | 0.05 |
ENST00000593360.1
|
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr3_+_49058444 | 0.05 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr11_-_45940343 | 0.04 |
ENST00000532681.1
|
PEX16
|
peroxisomal biogenesis factor 16 |
| chr3_-_149470229 | 0.04 |
ENST00000473414.1
|
COMMD2
|
COMM domain containing 2 |
| chr5_+_175740083 | 0.04 |
ENST00000332772.4
|
SIMC1
|
SUMO-interacting motifs containing 1 |
| chr17_-_4643161 | 0.04 |
ENST00000574412.1
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr4_-_54232144 | 0.03 |
ENST00000388940.4
ENST00000503450.1 ENST00000401642.3 |
SCFD2
|
sec1 family domain containing 2 |
| chr12_+_53693812 | 0.03 |
ENST00000549488.1
|
C12orf10
|
chromosome 12 open reading frame 10 |
| chr12_-_56321397 | 0.02 |
ENST00000557259.1
ENST00000549939.1 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr15_+_93443419 | 0.02 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr17_+_4643337 | 0.01 |
ENST00000592813.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr2_-_70780770 | 0.01 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr15_-_60683326 | 0.01 |
ENST00000559350.1
ENST00000558986.1 ENST00000560389.1 |
ANXA2
|
annexin A2 |
| chr15_-_49102781 | 0.00 |
ENST00000558591.1
|
CEP152
|
centrosomal protein 152kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of DMBX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.2 | 0.7 | GO:0018277 | protein deamination(GO:0018277) |
| 0.1 | 0.2 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.1 | GO:0072684 | mitochondrial tRNA 3'-trailer cleavage, endonucleolytic(GO:0072684) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.0 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.3 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.2 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.1 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.3 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.7 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.2 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.0 | GO:0032184 | SUMO polymer binding(GO:0032184) |