Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
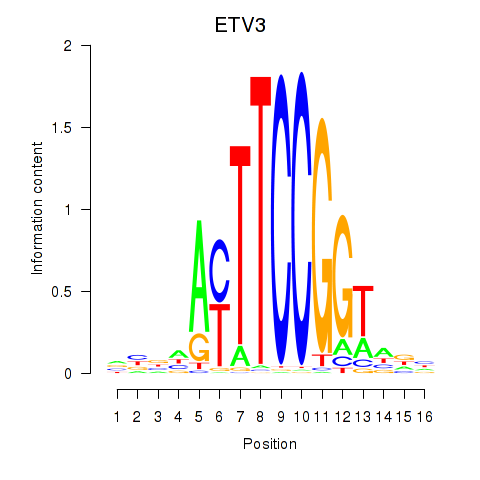
Results for ETV3
Z-value: 0.35
Transcription factors associated with ETV3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV3
|
ENSG00000117036.7 | ETS variant transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ETV3 | hg19_v2_chr1_-_157108130_157108173 | 0.58 | 7.2e-03 | Click! |
Activity profile of ETV3 motif
Sorted Z-values of ETV3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_118122996 | 0.65 |
ENST00000525386.1
ENST00000527472.1 ENST00000278949.4 |
MPZL3
|
myelin protein zero-like 3 |
| chr13_-_46626847 | 0.59 |
ENST00000242848.4
ENST00000282007.3 |
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr2_+_113885138 | 0.58 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr6_-_28303901 | 0.51 |
ENST00000439158.1
ENST00000446474.1 ENST00000414431.1 ENST00000344279.6 ENST00000453745.1 |
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr1_-_38455650 | 0.43 |
ENST00000448721.2
|
SF3A3
|
splicing factor 3a, subunit 3, 60kDa |
| chr13_-_46626820 | 0.43 |
ENST00000428921.1
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr18_+_72265084 | 0.41 |
ENST00000582337.1
|
ZNF407
|
zinc finger protein 407 |
| chr2_-_175260368 | 0.39 |
ENST00000342016.3
ENST00000362053.5 |
CIR1
|
corepressor interacting with RBPJ, 1 |
| chr1_+_38478432 | 0.39 |
ENST00000537711.1
|
UTP11L
|
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
| chr17_-_39093672 | 0.38 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr1_-_109618566 | 0.35 |
ENST00000338366.5
|
TAF13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr7_-_143991230 | 0.34 |
ENST00000543357.1
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35 |
| chr21_+_18885430 | 0.34 |
ENST00000356275.6
ENST00000400165.1 ENST00000400169.1 ENST00000306618.10 |
CXADR
|
coxsackie virus and adenovirus receptor |
| chr5_-_138862326 | 0.33 |
ENST00000330794.4
|
TMEM173
|
transmembrane protein 173 |
| chr2_+_175260451 | 0.33 |
ENST00000458563.1
ENST00000409673.3 ENST00000272732.6 ENST00000435964.1 |
SCRN3
|
secernin 3 |
| chr17_-_45266542 | 0.32 |
ENST00000531206.1
ENST00000527547.1 ENST00000446365.2 ENST00000575483.1 ENST00000066544.3 |
CDC27
|
cell division cycle 27 |
| chr7_+_139025105 | 0.32 |
ENST00000541170.3
|
C7orf55-LUC7L2
|
C7orf55-LUC7L2 readthrough |
| chr19_-_51522955 | 0.32 |
ENST00000358789.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr16_+_23652773 | 0.32 |
ENST00000563998.1
ENST00000568589.1 ENST00000568272.1 |
DCTN5
|
dynactin 5 (p25) |
| chr9_+_131452239 | 0.32 |
ENST00000372688.4
ENST00000372686.5 |
SET
|
SET nuclear oncogene |
| chr18_+_21033239 | 0.31 |
ENST00000581585.1
ENST00000577501.1 |
RIOK3
|
RIO kinase 3 |
| chr2_+_175260514 | 0.31 |
ENST00000424069.1
ENST00000427038.1 |
SCRN3
|
secernin 3 |
| chr6_-_28304152 | 0.31 |
ENST00000435857.1
|
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr11_-_116658695 | 0.31 |
ENST00000429220.1
ENST00000444935.1 |
ZNF259
|
zinc finger protein 259 |
| chr1_+_169337172 | 0.31 |
ENST00000367807.3
ENST00000367808.3 ENST00000329281.2 ENST00000420531.1 |
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr17_-_56065484 | 0.31 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr19_-_51523412 | 0.29 |
ENST00000391805.1
ENST00000599077.1 |
KLK10
|
kallikrein-related peptidase 10 |
| chr11_-_116658758 | 0.29 |
ENST00000227322.3
|
ZNF259
|
zinc finger protein 259 |
| chr4_-_100815525 | 0.29 |
ENST00000226522.8
ENST00000499666.2 |
LAMTOR3
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
| chr6_-_27440460 | 0.28 |
ENST00000377419.1
|
ZNF184
|
zinc finger protein 184 |
| chr1_+_10003486 | 0.28 |
ENST00000403197.1
ENST00000377205.1 |
NMNAT1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr22_-_36925124 | 0.28 |
ENST00000457241.1
|
EIF3D
|
eukaryotic translation initiation factor 3, subunit D |
| chr1_+_40506392 | 0.28 |
ENST00000414893.1
ENST00000414281.1 ENST00000420216.1 ENST00000372792.2 ENST00000372798.1 ENST00000340450.3 ENST00000372805.3 ENST00000435719.1 ENST00000427843.1 ENST00000417287.1 ENST00000424977.1 ENST00000446031.1 |
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr5_-_74062867 | 0.28 |
ENST00000509097.1
|
GFM2
|
G elongation factor, mitochondrial 2 |
| chr17_+_7387677 | 0.27 |
ENST00000322644.6
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr5_+_158690089 | 0.27 |
ENST00000296786.6
|
UBLCP1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr18_+_20513782 | 0.27 |
ENST00000399722.2
ENST00000399725.2 ENST00000399721.2 ENST00000583594.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr6_-_27440837 | 0.26 |
ENST00000211936.6
|
ZNF184
|
zinc finger protein 184 |
| chr21_-_33984456 | 0.26 |
ENST00000431216.1
ENST00000553001.1 ENST00000440966.1 |
AP000275.65
C21orf59
|
Uncharacterized protein chromosome 21 open reading frame 59 |
| chr1_+_169337412 | 0.26 |
ENST00000426663.1
|
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr1_+_40506255 | 0.26 |
ENST00000421589.1
|
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr3_+_179065474 | 0.26 |
ENST00000471841.1
ENST00000280653.7 |
MFN1
|
mitofusin 1 |
| chr11_+_131781290 | 0.26 |
ENST00000425719.2
ENST00000374784.1 |
NTM
|
neurotrimin |
| chr2_+_65454926 | 0.25 |
ENST00000542850.1
ENST00000377982.4 |
ACTR2
|
ARP2 actin-related protein 2 homolog (yeast) |
| chr1_+_28099700 | 0.25 |
ENST00000440806.2
|
STX12
|
syntaxin 12 |
| chr6_-_33239712 | 0.25 |
ENST00000436044.2
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr5_-_132202329 | 0.25 |
ENST00000378673.2
|
GDF9
|
growth differentiation factor 9 |
| chr2_-_230787879 | 0.25 |
ENST00000435716.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr11_+_118230287 | 0.24 |
ENST00000252108.3
ENST00000431736.2 |
UBE4A
|
ubiquitination factor E4A |
| chr6_+_28048753 | 0.24 |
ENST00000377325.1
|
ZNF165
|
zinc finger protein 165 |
| chr18_+_20513278 | 0.24 |
ENST00000327155.5
|
RBBP8
|
retinoblastoma binding protein 8 |
| chr3_+_15468862 | 0.24 |
ENST00000396842.2
|
EAF1
|
ELL associated factor 1 |
| chr3_+_15469058 | 0.24 |
ENST00000432764.2
|
EAF1
|
ELL associated factor 1 |
| chr7_+_99613212 | 0.24 |
ENST00000426572.1
ENST00000535170.1 |
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr4_+_56719782 | 0.24 |
ENST00000381295.2
ENST00000346134.7 ENST00000349598.6 |
EXOC1
|
exocyst complex component 1 |
| chr2_-_44588893 | 0.24 |
ENST00000409272.1
ENST00000410081.1 ENST00000541738.1 |
PREPL
|
prolyl endopeptidase-like |
| chr7_+_128379449 | 0.23 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr19_-_51523275 | 0.23 |
ENST00000309958.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr8_-_42698292 | 0.23 |
ENST00000529779.1
|
THAP1
|
THAP domain containing, apoptosis associated protein 1 |
| chr12_-_51566562 | 0.23 |
ENST00000548108.1
|
TFCP2
|
transcription factor CP2 |
| chr21_-_36260980 | 0.23 |
ENST00000344691.4
ENST00000358356.5 |
RUNX1
|
runt-related transcription factor 1 |
| chr5_-_138861926 | 0.23 |
ENST00000510817.1
|
TMEM173
|
transmembrane protein 173 |
| chr1_+_38478378 | 0.22 |
ENST00000373014.4
|
UTP11L
|
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
| chr2_+_187350973 | 0.22 |
ENST00000544130.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr11_-_17229480 | 0.22 |
ENST00000532035.1
ENST00000540361.1 |
PIK3C2A
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr6_-_149969871 | 0.22 |
ENST00000335643.8
ENST00000444282.1 |
KATNA1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr3_+_88199099 | 0.22 |
ENST00000486971.1
|
C3orf38
|
chromosome 3 open reading frame 38 |
| chr2_-_44588624 | 0.22 |
ENST00000438314.1
ENST00000409936.1 |
PREPL
|
prolyl endopeptidase-like |
| chr17_+_8316442 | 0.22 |
ENST00000582812.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr1_-_28415075 | 0.22 |
ENST00000373863.3
ENST00000436342.2 ENST00000540618.1 ENST00000545175.1 |
EYA3
|
eyes absent homolog 3 (Drosophila) |
| chr15_-_43398274 | 0.22 |
ENST00000382177.2
ENST00000290650.4 |
UBR1
|
ubiquitin protein ligase E3 component n-recognin 1 |
| chr22_-_38349552 | 0.22 |
ENST00000422191.1
ENST00000249079.2 ENST00000418863.1 ENST00000403305.1 ENST00000403026.1 |
C22orf23
|
chromosome 22 open reading frame 23 |
| chr10_-_50747064 | 0.22 |
ENST00000355832.5
ENST00000603152.1 ENST00000447839.2 |
ERCC6
PGBD3
ERCC6-PGBD3
|
excision repair cross-complementing rodent repair deficiency, complementation group 6 piggyBac transposable element derived 3 ERCC6-PGBD3 readthrough |
| chr15_+_74833518 | 0.21 |
ENST00000346246.5
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr22_-_30752861 | 0.21 |
ENST00000439242.1
ENST00000215793.8 |
SF3A1
|
splicing factor 3a, subunit 1, 120kDa |
| chr2_+_162016827 | 0.21 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr2_+_187350883 | 0.21 |
ENST00000337859.6
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr16_+_47495225 | 0.21 |
ENST00000299167.8
ENST00000323584.5 ENST00000563376.1 |
PHKB
|
phosphorylase kinase, beta |
| chr18_+_21032781 | 0.21 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr19_+_58095501 | 0.20 |
ENST00000536878.2
ENST00000597850.1 ENST00000597219.1 ENST00000598689.1 ENST00000599456.1 ENST00000307468.4 |
ZIK1
|
zinc finger protein interacting with K protein 1 |
| chr8_-_56685859 | 0.20 |
ENST00000523423.1
ENST00000523073.1 ENST00000519784.1 ENST00000434581.2 ENST00000519780.1 ENST00000521229.1 ENST00000522576.1 ENST00000523180.1 ENST00000522090.1 |
TMEM68
|
transmembrane protein 68 |
| chr2_-_44588679 | 0.20 |
ENST00000409411.1
|
PREPL
|
prolyl endopeptidase-like |
| chr2_-_44588694 | 0.20 |
ENST00000409957.1
|
PREPL
|
prolyl endopeptidase-like |
| chr2_-_175351744 | 0.20 |
ENST00000295500.4
ENST00000392552.2 ENST00000392551.2 |
GPR155
|
G protein-coupled receptor 155 |
| chr16_-_12009833 | 0.20 |
ENST00000420576.2
|
GSPT1
|
G1 to S phase transition 1 |
| chr15_-_44116873 | 0.20 |
ENST00000267812.3
|
MFAP1
|
microfibrillar-associated protein 1 |
| chr6_+_111303218 | 0.20 |
ENST00000441448.2
|
RPF2
|
ribosome production factor 2 homolog (S. cerevisiae) |
| chr19_-_37406923 | 0.20 |
ENST00000520965.1
|
ZNF829
|
zinc finger protein 829 |
| chr5_+_133984462 | 0.20 |
ENST00000398844.2
ENST00000322887.4 |
SEC24A
|
SEC24 family member A |
| chr3_+_28390637 | 0.20 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr16_+_47495201 | 0.19 |
ENST00000566044.1
ENST00000455779.1 |
PHKB
|
phosphorylase kinase, beta |
| chr18_+_33552667 | 0.19 |
ENST00000333234.5
|
C18orf21
|
chromosome 18 open reading frame 21 |
| chr1_+_93811438 | 0.19 |
ENST00000370272.4
ENST00000370267.1 |
DR1
|
down-regulator of transcription 1, TBP-binding (negative cofactor 2) |
| chr8_-_95565673 | 0.19 |
ENST00000437199.1
ENST00000297591.5 ENST00000421249.2 |
KIAA1429
|
KIAA1429 |
| chr2_+_65454863 | 0.19 |
ENST00000260641.5
|
ACTR2
|
ARP2 actin-related protein 2 homolog (yeast) |
| chr11_+_10772847 | 0.19 |
ENST00000524523.1
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr1_+_28655505 | 0.19 |
ENST00000373842.4
ENST00000398997.2 |
MED18
|
mediator complex subunit 18 |
| chr5_+_86564739 | 0.19 |
ENST00000456692.2
ENST00000512763.1 ENST00000506290.1 |
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1 |
| chr2_+_162016804 | 0.19 |
ENST00000392749.2
ENST00000440506.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr21_+_18885318 | 0.19 |
ENST00000400166.1
|
CXADR
|
coxsackie virus and adenovirus receptor |
| chr16_-_20817857 | 0.19 |
ENST00000563117.1
|
ERI2
|
ERI1 exoribonuclease family member 2 |
| chr11_-_57102947 | 0.19 |
ENST00000526696.1
|
SSRP1
|
structure specific recognition protein 1 |
| chr20_-_34638841 | 0.19 |
ENST00000565493.1
|
LINC00657
|
long intergenic non-protein coding RNA 657 |
| chr19_+_37407351 | 0.18 |
ENST00000455427.2
ENST00000587857.1 |
ZNF568
|
zinc finger protein 568 |
| chr4_-_83812248 | 0.18 |
ENST00000514326.1
ENST00000505434.1 ENST00000503058.1 ENST00000348405.4 ENST00000505984.1 ENST00000513858.1 ENST00000508479.1 ENST00000443462.2 ENST00000508502.1 ENST00000509142.1 ENST00000432794.1 ENST00000448323.1 ENST00000326950.5 ENST00000311785.7 |
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr2_+_162016916 | 0.18 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr5_-_102455801 | 0.18 |
ENST00000508629.1
ENST00000399004.2 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chr14_+_24701870 | 0.18 |
ENST00000561035.1
ENST00000559409.1 ENST00000558865.1 ENST00000558279.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr16_-_47495170 | 0.18 |
ENST00000320640.6
ENST00000544001.2 |
ITFG1
|
integrin alpha FG-GAP repeat containing 1 |
| chr1_+_28099683 | 0.18 |
ENST00000373943.4
|
STX12
|
syntaxin 12 |
| chr2_-_230786032 | 0.18 |
ENST00000428959.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr3_+_113465866 | 0.18 |
ENST00000273398.3
ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr3_+_47324424 | 0.18 |
ENST00000437353.1
ENST00000232766.5 ENST00000455924.2 |
KLHL18
|
kelch-like family member 18 |
| chr6_-_33239612 | 0.18 |
ENST00000482399.1
ENST00000445902.2 |
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr8_-_53626974 | 0.18 |
ENST00000435644.2
ENST00000518710.1 ENST00000025008.5 ENST00000517963.1 |
RB1CC1
|
RB1-inducible coiled-coil 1 |
| chr14_-_24711764 | 0.18 |
ENST00000557921.1
ENST00000558476.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr5_+_82373379 | 0.18 |
ENST00000396027.4
ENST00000511817.1 |
XRCC4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr4_-_83812402 | 0.18 |
ENST00000395310.2
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr13_-_108867846 | 0.18 |
ENST00000442234.1
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr1_+_63989004 | 0.18 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr1_+_110091189 | 0.18 |
ENST00000369851.4
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr11_-_125773085 | 0.17 |
ENST00000227474.3
ENST00000534158.1 ENST00000529801.1 |
PUS3
|
pseudouridylate synthase 3 |
| chr1_+_203765437 | 0.17 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr20_+_49126881 | 0.17 |
ENST00000371621.3
ENST00000541713.1 |
PTPN1
|
protein tyrosine phosphatase, non-receptor type 1 |
| chr5_-_74062930 | 0.17 |
ENST00000509430.1
ENST00000345239.2 ENST00000427854.2 ENST00000506778.1 |
GFM2
|
G elongation factor, mitochondrial 2 |
| chr2_-_201753717 | 0.17 |
ENST00000409264.2
|
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr3_+_88198838 | 0.17 |
ENST00000318887.3
|
C3orf38
|
chromosome 3 open reading frame 38 |
| chr12_-_75905374 | 0.17 |
ENST00000438169.2
ENST00000229214.4 |
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr11_+_10772534 | 0.17 |
ENST00000361367.2
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr4_-_76439483 | 0.17 |
ENST00000380840.2
ENST00000513257.1 ENST00000507014.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr7_-_108209897 | 0.17 |
ENST00000313516.5
|
THAP5
|
THAP domain containing 5 |
| chr20_+_43595115 | 0.17 |
ENST00000372806.3
ENST00000396731.4 ENST00000372801.1 ENST00000499879.2 |
STK4
|
serine/threonine kinase 4 |
| chr12_+_109490370 | 0.17 |
ENST00000257548.5
ENST00000536723.1 ENST00000536393.1 |
USP30
|
ubiquitin specific peptidase 30 |
| chr7_-_91509986 | 0.17 |
ENST00000456229.1
ENST00000442961.1 ENST00000406735.2 ENST00000419292.1 ENST00000351870.3 |
MTERF
|
mitochondrial transcription termination factor |
| chr5_+_125758865 | 0.17 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr5_+_82373317 | 0.16 |
ENST00000282268.3
ENST00000338635.6 |
XRCC4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr10_-_14996017 | 0.16 |
ENST00000378241.1
ENST00000456122.1 ENST00000418843.1 ENST00000378249.1 ENST00000396817.2 ENST00000378255.1 ENST00000378254.1 ENST00000378278.2 ENST00000357717.2 |
DCLRE1C
|
DNA cross-link repair 1C |
| chr18_-_47808120 | 0.16 |
ENST00000587605.1
ENST00000591416.1 ENST00000382948.5 ENST00000269471.5 ENST00000269468.5 ENST00000347968.3 ENST00000436910.1 ENST00000349085.2 |
MBD1
|
methyl-CpG binding domain protein 1 |
| chr14_-_81687575 | 0.16 |
ENST00000434192.2
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa |
| chr8_-_38034234 | 0.16 |
ENST00000311351.4
|
LSM1
|
LSM1 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr21_-_33985127 | 0.16 |
ENST00000290155.3
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr6_-_26659913 | 0.16 |
ENST00000480036.1
ENST00000415922.2 |
ZNF322
|
zinc finger protein 322 |
| chr11_+_63606477 | 0.16 |
ENST00000508192.1
ENST00000361128.5 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr2_-_69870747 | 0.16 |
ENST00000409068.1
|
AAK1
|
AP2 associated kinase 1 |
| chr17_-_65362678 | 0.16 |
ENST00000357146.4
ENST00000356126.3 |
PSMD12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 |
| chr2_-_110371777 | 0.15 |
ENST00000397712.2
|
SEPT10
|
septin 10 |
| chr12_-_49110613 | 0.15 |
ENST00000261900.3
|
CCNT1
|
cyclin T1 |
| chr8_+_56685701 | 0.15 |
ENST00000260129.5
|
TGS1
|
trimethylguanosine synthase 1 |
| chr6_+_4021554 | 0.15 |
ENST00000337659.6
|
PRPF4B
|
pre-mRNA processing factor 4B |
| chr14_+_90722839 | 0.15 |
ENST00000261303.8
ENST00000553835.1 |
PSMC1
|
proteasome (prosome, macropain) 26S subunit, ATPase, 1 |
| chr16_+_19535133 | 0.15 |
ENST00000396212.2
ENST00000381396.5 |
CCP110
|
centriolar coiled coil protein 110kDa |
| chr10_+_121652204 | 0.15 |
ENST00000369075.3
ENST00000543134.1 |
SEC23IP
|
SEC23 interacting protein |
| chr4_-_77069533 | 0.15 |
ENST00000514987.1
ENST00000458189.2 ENST00000514901.1 ENST00000342467.6 |
NUP54
|
nucleoporin 54kDa |
| chr22_-_36877371 | 0.15 |
ENST00000403313.1
|
TXN2
|
thioredoxin 2 |
| chr5_-_102898465 | 0.15 |
ENST00000507423.1
ENST00000230792.2 |
NUDT12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr8_-_42698433 | 0.15 |
ENST00000345117.2
ENST00000254250.3 |
THAP1
|
THAP domain containing, apoptosis associated protein 1 |
| chr16_-_3493528 | 0.15 |
ENST00000301744.4
|
ZNF597
|
zinc finger protein 597 |
| chr1_+_100315613 | 0.15 |
ENST00000361915.3
|
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr1_-_52344416 | 0.15 |
ENST00000544028.1
|
NRD1
|
nardilysin (N-arginine dibasic convertase) |
| chr8_+_146052849 | 0.14 |
ENST00000532777.1
ENST00000325241.6 ENST00000446747.2 ENST00000525266.1 ENST00000544249.1 ENST00000325217.5 ENST00000533314.1 ENST00000527218.1 ENST00000529819.1 ENST00000528372.1 |
ZNF7
|
zinc finger protein 7 |
| chr7_+_128379346 | 0.14 |
ENST00000535011.2
ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU
|
calumenin |
| chr3_+_108308559 | 0.14 |
ENST00000486815.1
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr1_-_115323245 | 0.14 |
ENST00000060969.5
ENST00000369528.5 |
SIKE1
|
suppressor of IKBKE 1 |
| chr22_+_38245414 | 0.14 |
ENST00000381683.6
ENST00000414316.1 ENST00000406934.1 ENST00000451427.1 |
EIF3L
|
eukaryotic translation initiation factor 3, subunit L |
| chr2_-_230786378 | 0.14 |
ENST00000430954.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr3_-_139108475 | 0.14 |
ENST00000515006.1
ENST00000513274.1 ENST00000514508.1 ENST00000507777.1 ENST00000512153.1 ENST00000333188.5 |
COPB2
|
coatomer protein complex, subunit beta 2 (beta prime) |
| chr16_-_20817753 | 0.14 |
ENST00000389345.5
ENST00000300005.3 ENST00000357967.4 ENST00000569729.1 |
ERI2
|
ERI1 exoribonuclease family member 2 |
| chr5_+_130506475 | 0.14 |
ENST00000379380.4
|
LYRM7
|
LYR motif containing 7 |
| chr18_-_29522989 | 0.14 |
ENST00000582539.1
ENST00000283351.4 ENST00000582513.1 |
TRAPPC8
|
trafficking protein particle complex 8 |
| chr7_+_99613195 | 0.14 |
ENST00000324306.6
|
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr2_-_110371720 | 0.14 |
ENST00000356688.4
|
SEPT10
|
septin 10 |
| chr19_+_37407212 | 0.14 |
ENST00000427117.1
ENST00000587130.1 ENST00000333987.7 ENST00000415168.1 ENST00000444991.1 |
ZNF568
|
zinc finger protein 568 |
| chr2_-_201753859 | 0.14 |
ENST00000409361.1
ENST00000392283.4 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr3_-_88198965 | 0.14 |
ENST00000467332.1
ENST00000462901.1 |
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr22_-_19466454 | 0.14 |
ENST00000494054.1
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr3_-_47324079 | 0.14 |
ENST00000352910.4
|
KIF9
|
kinesin family member 9 |
| chrX_+_70586082 | 0.14 |
ENST00000373790.4
ENST00000449580.1 ENST00000423759.1 |
TAF1
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr12_-_51566592 | 0.14 |
ENST00000257915.5
ENST00000548115.1 |
TFCP2
|
transcription factor CP2 |
| chr5_-_82373260 | 0.14 |
ENST00000502346.1
|
TMEM167A
|
transmembrane protein 167A |
| chr20_+_44486246 | 0.14 |
ENST00000255152.2
ENST00000454862.2 |
ZSWIM3
|
zinc finger, SWIM-type containing 3 |
| chr2_+_170440902 | 0.14 |
ENST00000448752.2
ENST00000418888.1 ENST00000414307.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr14_-_24711470 | 0.14 |
ENST00000559969.1
|
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr3_+_179322481 | 0.14 |
ENST00000259037.3
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr14_-_102553371 | 0.14 |
ENST00000553585.1
ENST00000216281.8 |
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr5_+_125758813 | 0.13 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr13_+_100153665 | 0.13 |
ENST00000376387.4
|
TM9SF2
|
transmembrane 9 superfamily member 2 |
| chr14_-_77843390 | 0.13 |
ENST00000216468.7
|
TMED8
|
transmembrane emp24 protein transport domain containing 8 |
| chr2_-_73964447 | 0.13 |
ENST00000272424.5
ENST00000409716.2 ENST00000318190.7 |
TPRKB
|
TP53RK binding protein |
| chr15_-_40331356 | 0.13 |
ENST00000560773.1
ENST00000267884.6 |
SRP14
|
signal recognition particle 14kDa (homologous Alu RNA binding protein) |
| chr17_-_10600818 | 0.13 |
ENST00000577427.1
ENST00000255390.5 |
SCO1
|
SCO1 cytochrome c oxidase assembly protein |
| chr6_-_86303523 | 0.13 |
ENST00000513865.1
ENST00000369627.2 ENST00000514419.1 ENST00000509338.1 ENST00000314673.3 ENST00000346348.3 |
SNX14
|
sorting nexin 14 |
| chr5_+_79703823 | 0.13 |
ENST00000338008.5
ENST00000510158.1 ENST00000505560.1 |
ZFYVE16
|
zinc finger, FYVE domain containing 16 |
| chrX_+_70586140 | 0.13 |
ENST00000276072.3
|
TAF1
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr6_-_149969829 | 0.13 |
ENST00000367411.2
|
KATNA1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr16_-_23652570 | 0.13 |
ENST00000261584.4
|
PALB2
|
partner and localizer of BRCA2 |
| chr12_+_21590549 | 0.13 |
ENST00000545178.1
ENST00000240651.9 |
PYROXD1
|
pyridine nucleotide-disulphide oxidoreductase domain 1 |
| chr11_+_63606558 | 0.13 |
ENST00000350490.7
ENST00000502399.3 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr1_+_168148273 | 0.13 |
ENST00000367830.3
|
TIPRL
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr2_+_44589036 | 0.13 |
ENST00000402247.1
ENST00000407131.1 ENST00000403853.3 ENST00000378494.3 |
CAMKMT
|
calmodulin-lysine N-methyltransferase |
| chr4_-_155471528 | 0.13 |
ENST00000302078.5
ENST00000499023.2 |
PLRG1
|
pleiotropic regulator 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ETV3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 | 0.5 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.5 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.3 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.1 | 0.4 | GO:2001162 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.1 | 0.5 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.1 | 0.2 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.1 | 0.8 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 0.6 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.6 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.4 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 0.2 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.1 | 0.6 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.2 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 0.2 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 0.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.3 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.0 | 0.1 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.3 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.0 | 0.2 | GO:0035936 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.3 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.5 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.4 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.6 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.2 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.2 | GO:0036260 | 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.3 | GO:0075713 | establishment of integrated proviral latency(GO:0075713) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.2 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.0 | 0.1 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.0 | 0.1 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.0 | 0.1 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.1 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.3 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.4 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.2 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.2 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.2 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.2 | GO:0071484 | cellular response to light intensity(GO:0071484) |
| 0.0 | 0.0 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.0 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.0 | 0.9 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.0 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.7 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.1 | GO:0071964 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.0 | 0.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.0 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.5 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.5 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.5 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.1 | 0.2 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 0.6 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.2 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.5 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.0 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.2 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.3 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.3 | GO:0042588 | zymogen granule(GO:0042588) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 0.5 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.3 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.1 | 0.6 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.3 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.2 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.3 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.6 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.7 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.4 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.9 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.3 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.1 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.0 | 0.1 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.0 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.5 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.1 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |