Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
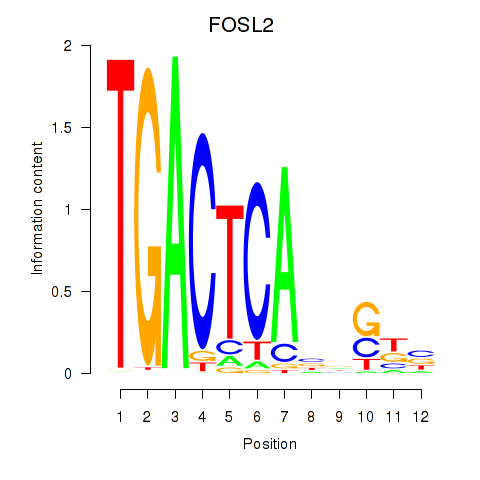
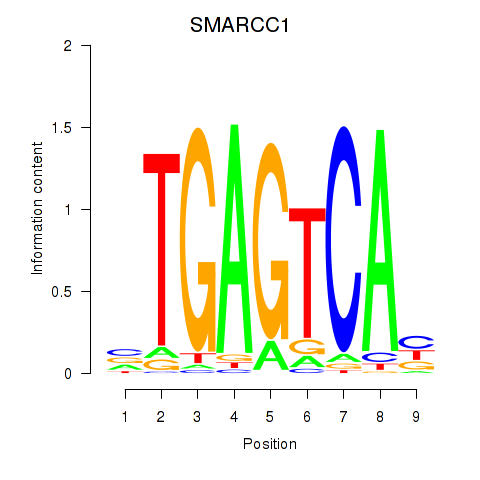
Results for FOSL2_SMARCC1
Z-value: 1.55
Transcription factors associated with FOSL2_SMARCC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOSL2
|
ENSG00000075426.7 | FOS like 2, AP-1 transcription factor subunit |
|
SMARCC1
|
ENSG00000173473.6 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SMARCC1 | hg19_v2_chr3_-_47823298_47823423 | 0.76 | 1.1e-04 | Click! |
| FOSL2 | hg19_v2_chr2_+_28615669_28615733 | 0.68 | 8.9e-04 | Click! |
Activity profile of FOSL2_SMARCC1 motif
Sorted Z-values of FOSL2_SMARCC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_39769005 | 23.28 |
ENST00000301653.4
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr18_+_21452804 | 18.23 |
ENST00000269217.6
|
LAMA3
|
laminin, alpha 3 |
| chr18_+_21452964 | 17.70 |
ENST00000587184.1
|
LAMA3
|
laminin, alpha 3 |
| chr2_+_113875466 | 14.34 |
ENST00000361779.3
ENST00000259206.5 ENST00000354115.2 |
IL1RN
|
interleukin 1 receptor antagonist |
| chr7_+_129932974 | 13.33 |
ENST00000445470.2
ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr3_-_151034734 | 9.03 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr7_+_73245193 | 8.73 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr1_-_156675368 | 8.21 |
ENST00000368222.3
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr17_-_39743139 | 7.84 |
ENST00000167586.6
|
KRT14
|
keratin 14 |
| chr6_-_113953705 | 7.66 |
ENST00000452675.1
|
RP11-367G18.1
|
RP11-367G18.1 |
| chr1_+_150480551 | 7.35 |
ENST00000369049.4
ENST00000369047.4 |
ECM1
|
extracellular matrix protein 1 |
| chr11_-_102668879 | 7.35 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr15_+_41136586 | 7.17 |
ENST00000431806.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr1_+_150480576 | 6.88 |
ENST00000346569.6
|
ECM1
|
extracellular matrix protein 1 |
| chr19_+_39279838 | 6.86 |
ENST00000314980.4
|
LGALS7B
|
lectin, galactoside-binding, soluble, 7B |
| chr2_+_169926047 | 6.80 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr1_+_223889310 | 6.72 |
ENST00000434648.1
|
CAPN2
|
calpain 2, (m/II) large subunit |
| chr15_+_41136216 | 6.66 |
ENST00000562057.1
ENST00000344051.4 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr6_+_41604747 | 6.50 |
ENST00000419164.1
ENST00000373051.2 |
MDFI
|
MyoD family inhibitor |
| chr2_+_208104351 | 6.40 |
ENST00000440326.1
|
AC007879.7
|
AC007879.7 |
| chr20_+_44637526 | 6.38 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr1_-_153538011 | 6.15 |
ENST00000368707.4
|
S100A2
|
S100 calcium binding protein A2 |
| chr18_+_61442629 | 6.13 |
ENST00000398019.2
ENST00000540675.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr5_+_149877334 | 5.99 |
ENST00000523767.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr19_-_36004543 | 5.80 |
ENST00000339686.3
ENST00000447113.2 ENST00000440396.1 |
DMKN
|
dermokine |
| chr2_+_208104497 | 5.78 |
ENST00000430494.1
|
AC007879.7
|
AC007879.7 |
| chr18_-_68004529 | 5.64 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr19_-_36019123 | 5.63 |
ENST00000588674.1
ENST00000452271.2 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr15_+_41136734 | 5.57 |
ENST00000568580.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chrX_+_115567767 | 5.51 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr5_+_150020240 | 5.46 |
ENST00000519664.1
|
SYNPO
|
synaptopodin |
| chr1_+_183155373 | 5.43 |
ENST00000493293.1
ENST00000264144.4 |
LAMC2
|
laminin, gamma 2 |
| chr1_+_201252580 | 5.30 |
ENST00000367324.3
ENST00000263946.3 |
PKP1
|
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) |
| chr19_-_51523412 | 5.29 |
ENST00000391805.1
ENST00000599077.1 |
KLK10
|
kallikrein-related peptidase 10 |
| chr12_-_95009837 | 5.22 |
ENST00000551457.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr3_-_12800751 | 5.11 |
ENST00000435218.2
ENST00000435575.1 |
TMEM40
|
transmembrane protein 40 |
| chr3_-_121379739 | 5.03 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr6_+_41604620 | 5.02 |
ENST00000432027.1
|
MDFI
|
MyoD family inhibitor |
| chr16_-_2908155 | 4.99 |
ENST00000571228.1
ENST00000161006.3 |
PRSS22
|
protease, serine, 22 |
| chr1_+_26606608 | 4.99 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr11_+_12308447 | 4.98 |
ENST00000256186.2
|
MICALCL
|
MICAL C-terminal like |
| chr17_+_7255208 | 4.96 |
ENST00000333751.3
|
KCTD11
|
potassium channel tetramerization domain containing 11 |
| chr1_-_153029980 | 4.92 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr8_+_120220561 | 4.88 |
ENST00000276681.6
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene) |
| chr13_+_78109884 | 4.77 |
ENST00000377246.3
ENST00000349847.3 |
SCEL
|
sciellin |
| chr15_+_98503922 | 4.70 |
ENST00000268042.6
|
ARRDC4
|
arrestin domain containing 4 |
| chr13_+_78109804 | 4.64 |
ENST00000535157.1
|
SCEL
|
sciellin |
| chr19_-_51523275 | 4.57 |
ENST00000309958.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr1_+_35220613 | 4.56 |
ENST00000338513.1
|
GJB5
|
gap junction protein, beta 5, 31.1kDa |
| chr19_-_35992780 | 4.46 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr1_-_43751230 | 4.44 |
ENST00000523677.1
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr15_+_41136369 | 4.37 |
ENST00000563656.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr1_+_174844645 | 4.31 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr21_-_28215332 | 4.29 |
ENST00000517777.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr11_-_118134997 | 4.27 |
ENST00000278937.2
|
MPZL2
|
myelin protein zero-like 2 |
| chr12_-_95010147 | 4.14 |
ENST00000548918.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr11_-_118135160 | 4.13 |
ENST00000438295.2
|
MPZL2
|
myelin protein zero-like 2 |
| chr4_-_77819002 | 4.12 |
ENST00000334306.2
|
SOWAHB
|
sosondowah ankyrin repeat domain family member B |
| chr12_-_52845910 | 4.07 |
ENST00000252252.3
|
KRT6B
|
keratin 6B |
| chr9_+_71820057 | 4.07 |
ENST00000539225.1
|
TJP2
|
tight junction protein 2 |
| chr20_-_1309809 | 4.06 |
ENST00000360779.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr19_-_51587502 | 4.02 |
ENST00000156499.2
ENST00000391802.1 |
KLK14
|
kallikrein-related peptidase 14 |
| chrX_-_48326764 | 3.98 |
ENST00000413668.1
ENST00000441948.1 |
SLC38A5
|
solute carrier family 38, member 5 |
| chr11_-_65429891 | 3.97 |
ENST00000527874.1
|
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr6_+_106959718 | 3.96 |
ENST00000369066.3
|
AIM1
|
absent in melanoma 1 |
| chr14_+_21510385 | 3.90 |
ENST00000298690.4
|
RNASE7
|
ribonuclease, RNase A family, 7 |
| chr1_-_117753540 | 3.84 |
ENST00000328189.3
ENST00000369458.3 |
VTCN1
|
V-set domain containing T cell activation inhibitor 1 |
| chr9_+_71819927 | 3.81 |
ENST00000535702.1
|
TJP2
|
tight junction protein 2 |
| chr1_-_159924006 | 3.80 |
ENST00000368092.3
ENST00000368093.3 |
SLAMF9
|
SLAM family member 9 |
| chr8_+_22844995 | 3.77 |
ENST00000524077.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr5_+_150020214 | 3.77 |
ENST00000307662.4
|
SYNPO
|
synaptopodin |
| chr8_+_144821557 | 3.76 |
ENST00000534398.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr2_-_113594279 | 3.62 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr1_-_28520384 | 3.61 |
ENST00000305392.3
|
PTAFR
|
platelet-activating factor receptor |
| chr19_-_51472823 | 3.57 |
ENST00000310157.2
|
KLK6
|
kallikrein-related peptidase 6 |
| chr6_+_30853002 | 3.51 |
ENST00000421124.2
ENST00000512725.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr13_-_86373536 | 3.51 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr16_+_57662138 | 3.49 |
ENST00000562414.1
ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr19_-_39264072 | 3.49 |
ENST00000599035.1
ENST00000378626.4 |
LGALS7
|
lectin, galactoside-binding, soluble, 7 |
| chr3_+_184038234 | 3.49 |
ENST00000427607.1
ENST00000457456.1 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_-_28520447 | 3.39 |
ENST00000539896.1
|
PTAFR
|
platelet-activating factor receptor |
| chr14_-_24806588 | 3.35 |
ENST00000555591.1
ENST00000554569.1 |
RP11-934B9.3
RIPK3
|
Uncharacterized protein receptor-interacting serine-threonine kinase 3 |
| chr12_+_13349650 | 3.34 |
ENST00000256951.5
ENST00000431267.2 ENST00000542474.1 ENST00000544053.1 |
EMP1
|
epithelial membrane protein 1 |
| chr2_-_166651152 | 3.30 |
ENST00000431484.1
ENST00000412248.1 |
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) |
| chr3_-_69402828 | 3.26 |
ENST00000460709.1
|
FRMD4B
|
FERM domain containing 4B |
| chr2_-_113542063 | 3.25 |
ENST00000263339.3
|
IL1A
|
interleukin 1, alpha |
| chr14_-_75083313 | 3.24 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr11_-_6341844 | 3.22 |
ENST00000303927.3
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr9_+_34992846 | 3.19 |
ENST00000443266.1
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr17_-_7493390 | 3.19 |
ENST00000538513.2
ENST00000570788.1 ENST00000250055.2 |
SOX15
|
SRY (sex determining region Y)-box 15 |
| chr2_-_166651191 | 3.18 |
ENST00000392701.3
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) |
| chr17_+_48611853 | 3.18 |
ENST00000507709.1
ENST00000515126.1 ENST00000507467.1 |
EPN3
|
epsin 3 |
| chr6_-_35888824 | 3.17 |
ENST00000361690.3
ENST00000512445.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr6_+_47666275 | 3.17 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr9_-_114246332 | 3.10 |
ENST00000602978.1
|
KIAA0368
|
KIAA0368 |
| chr3_+_184037466 | 3.07 |
ENST00000441154.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr18_-_74207146 | 3.00 |
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516 |
| chrX_+_99899180 | 2.99 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr6_+_292051 | 2.99 |
ENST00000344450.5
|
DUSP22
|
dual specificity phosphatase 22 |
| chr6_+_30852738 | 2.99 |
ENST00000508312.1
ENST00000512336.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr12_+_69202795 | 2.97 |
ENST00000539479.1
ENST00000393415.3 ENST00000523991.1 ENST00000543323.1 ENST00000393416.2 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr1_+_154377669 | 2.92 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr7_+_55177416 | 2.88 |
ENST00000450046.1
ENST00000454757.2 |
EGFR
|
epidermal growth factor receptor |
| chr2_+_47596634 | 2.88 |
ENST00000419334.1
|
EPCAM
|
epithelial cell adhesion molecule |
| chr1_-_9953295 | 2.86 |
ENST00000377258.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr1_-_153113927 | 2.84 |
ENST00000368752.4
|
SPRR2B
|
small proline-rich protein 2B |
| chr11_+_101983176 | 2.84 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr9_-_35112376 | 2.83 |
ENST00000488109.2
|
FAM214B
|
family with sequence similarity 214, member B |
| chr10_+_75668916 | 2.78 |
ENST00000481390.1
|
PLAU
|
plasminogen activator, urokinase |
| chr10_+_24755416 | 2.77 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr11_-_66103932 | 2.77 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr2_+_169923504 | 2.73 |
ENST00000357546.2
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr18_+_61445007 | 2.72 |
ENST00000447428.1
ENST00000546027.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr18_+_61254570 | 2.70 |
ENST00000344731.5
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr4_-_74088800 | 2.69 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr13_-_41593425 | 2.66 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr1_+_27189631 | 2.65 |
ENST00000339276.4
|
SFN
|
stratifin |
| chr17_+_7482785 | 2.64 |
ENST00000250092.6
ENST00000380498.6 ENST00000584502.1 |
CD68
|
CD68 molecule |
| chr7_-_20256965 | 2.62 |
ENST00000400331.5
ENST00000332878.4 |
MACC1
|
metastasis associated in colon cancer 1 |
| chr20_+_4667094 | 2.61 |
ENST00000424424.1
ENST00000457586.1 |
PRNP
|
prion protein |
| chr12_-_48152853 | 2.59 |
ENST00000171000.4
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr16_+_57662527 | 2.54 |
ENST00000563374.1
ENST00000568234.1 ENST00000565770.1 ENST00000564338.1 ENST00000566164.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr1_-_209824643 | 2.52 |
ENST00000391911.1
ENST00000415782.1 |
LAMB3
|
laminin, beta 3 |
| chr6_-_56507586 | 2.51 |
ENST00000439203.1
ENST00000518935.1 ENST00000446842.2 ENST00000370765.6 ENST00000244364.6 |
DST
|
dystonin |
| chr1_-_27816641 | 2.51 |
ENST00000430629.2
|
WASF2
|
WAS protein family, member 2 |
| chr8_+_22844913 | 2.49 |
ENST00000519685.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr1_+_26605618 | 2.45 |
ENST00000270792.5
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr1_+_36621174 | 2.43 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr9_-_73029540 | 2.41 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr8_+_125860939 | 2.40 |
ENST00000525292.1
ENST00000528090.1 |
LINC00964
|
long intergenic non-protein coding RNA 964 |
| chr17_+_75446629 | 2.39 |
ENST00000588958.2
ENST00000586128.1 |
SEPT9
|
septin 9 |
| chr18_+_34124507 | 2.38 |
ENST00000591635.1
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr11_-_65430251 | 2.38 |
ENST00000534283.1
ENST00000527749.1 ENST00000533187.1 ENST00000525693.1 ENST00000534558.1 ENST00000532879.1 ENST00000532999.1 |
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr11_-_62323702 | 2.37 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr1_-_151965048 | 2.37 |
ENST00000368809.1
|
S100A10
|
S100 calcium binding protein A10 |
| chr18_+_61637159 | 2.32 |
ENST00000397985.2
ENST00000353706.2 ENST00000542677.1 ENST00000397988.3 ENST00000448851.1 |
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| chr15_+_41136263 | 2.32 |
ENST00000568823.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr3_+_184038073 | 2.32 |
ENST00000428387.1
ENST00000434061.2 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr6_+_74405804 | 2.31 |
ENST00000287097.5
|
CD109
|
CD109 molecule |
| chr5_-_41794663 | 2.30 |
ENST00000510634.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr5_+_149877440 | 2.29 |
ENST00000518299.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr14_-_91710852 | 2.28 |
ENST00000535815.1
ENST00000529102.1 |
GPR68
|
G protein-coupled receptor 68 |
| chr5_-_138533973 | 2.28 |
ENST00000507002.1
ENST00000505830.1 ENST00000508639.1 ENST00000265195.5 |
SIL1
|
SIL1 nucleotide exchange factor |
| chr15_+_40532058 | 2.26 |
ENST00000260404.4
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr14_+_71165292 | 2.22 |
ENST00000553682.1
|
RP6-65G23.1
|
RP6-65G23.1 |
| chr2_+_89952792 | 2.21 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
| chr2_+_169923577 | 2.19 |
ENST00000432060.2
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr18_+_61445205 | 2.18 |
ENST00000431370.1
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr1_-_6662919 | 2.16 |
ENST00000377658.4
ENST00000377663.3 |
KLHL21
|
kelch-like family member 21 |
| chr6_+_74405501 | 2.16 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr11_-_96076334 | 2.15 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr15_+_67420441 | 2.11 |
ENST00000558894.1
|
SMAD3
|
SMAD family member 3 |
| chr4_-_36246060 | 2.11 |
ENST00000303965.4
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr7_+_90338712 | 2.09 |
ENST00000265741.3
ENST00000406263.1 |
CDK14
|
cyclin-dependent kinase 14 |
| chr19_-_58485895 | 2.08 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr12_-_48152611 | 2.06 |
ENST00000389212.3
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr11_+_131240373 | 2.06 |
ENST00000374791.3
ENST00000436745.1 |
NTM
|
neurotrimin |
| chr17_-_1508379 | 2.05 |
ENST00000412517.3
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr4_+_3388057 | 2.05 |
ENST00000538395.1
|
RGS12
|
regulator of G-protein signaling 12 |
| chr3_-_99833333 | 2.04 |
ENST00000354552.3
ENST00000331335.5 ENST00000398326.2 |
FILIP1L
|
filamin A interacting protein 1-like |
| chr11_-_66104237 | 2.04 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr7_-_150721570 | 2.03 |
ENST00000377974.2
ENST00000444312.1 ENST00000605938.1 ENST00000605952.1 |
ATG9B
|
autophagy related 9B |
| chr11_-_94965667 | 2.03 |
ENST00000542176.1
ENST00000278499.2 |
SESN3
|
sestrin 3 |
| chr16_+_67571351 | 2.02 |
ENST00000428437.2
ENST00000569253.1 |
FAM65A
|
family with sequence similarity 65, member A |
| chr11_-_66103867 | 1.99 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr2_-_64371546 | 1.94 |
ENST00000358912.4
|
PELI1
|
pellino E3 ubiquitin protein ligase 1 |
| chr11_+_28724129 | 1.92 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr3_-_48598547 | 1.92 |
ENST00000536104.1
ENST00000452531.1 |
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr6_+_30852130 | 1.91 |
ENST00000428153.2
ENST00000376568.3 ENST00000452441.1 ENST00000515219.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr1_+_27669719 | 1.89 |
ENST00000473280.1
|
SYTL1
|
synaptotagmin-like 1 |
| chr1_-_89488510 | 1.89 |
ENST00000564665.1
ENST00000370481.4 |
GBP3
|
guanylate binding protein 3 |
| chr1_-_179112189 | 1.89 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr1_-_202777535 | 1.89 |
ENST00000367264.2
|
KDM5B
|
lysine (K)-specific demethylase 5B |
| chr4_+_159727222 | 1.86 |
ENST00000512986.1
|
FNIP2
|
folliculin interacting protein 2 |
| chr17_+_74381343 | 1.85 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr3_-_18466787 | 1.84 |
ENST00000338745.6
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr4_-_102268708 | 1.84 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr1_-_6659876 | 1.83 |
ENST00000496707.1
|
KLHL21
|
kelch-like family member 21 |
| chr7_+_143078379 | 1.82 |
ENST00000449630.1
ENST00000457235.1 |
ZYX
|
zyxin |
| chr1_+_901847 | 1.81 |
ENST00000379410.3
ENST00000379409.2 ENST00000379407.3 |
PLEKHN1
|
pleckstrin homology domain containing, family N member 1 |
| chr5_-_41794313 | 1.81 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr17_+_73717407 | 1.80 |
ENST00000579662.1
|
ITGB4
|
integrin, beta 4 |
| chr12_+_13349711 | 1.79 |
ENST00000538364.1
ENST00000396301.3 |
EMP1
|
epithelial membrane protein 1 |
| chr16_+_57662596 | 1.79 |
ENST00000567397.1
ENST00000568979.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr1_+_223889285 | 1.78 |
ENST00000433674.2
|
CAPN2
|
calpain 2, (m/II) large subunit |
| chr16_+_57662419 | 1.77 |
ENST00000388812.4
ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56
|
G protein-coupled receptor 56 |
| chr1_+_6511651 | 1.75 |
ENST00000434576.1
|
ESPN
|
espin |
| chr12_+_69202975 | 1.74 |
ENST00000544561.1
ENST00000393410.1 ENST00000299252.4 ENST00000360430.2 ENST00000517852.1 ENST00000545204.1 ENST00000393413.3 ENST00000350057.5 ENST00000348801.2 ENST00000478070.1 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr1_-_43751276 | 1.72 |
ENST00000423420.1
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr19_-_51568324 | 1.71 |
ENST00000595547.1
ENST00000335422.3 ENST00000595793.1 ENST00000596955.1 |
KLK13
|
kallikrein-related peptidase 13 |
| chr11_-_111794446 | 1.70 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr8_+_123875624 | 1.70 |
ENST00000534247.1
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr11_-_65667884 | 1.68 |
ENST00000448083.2
ENST00000531493.1 ENST00000532401.1 |
FOSL1
|
FOS-like antigen 1 |
| chr11_+_10476851 | 1.68 |
ENST00000396553.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr1_-_108231101 | 1.67 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr17_+_73717551 | 1.67 |
ENST00000450894.3
|
ITGB4
|
integrin, beta 4 |
| chr17_+_73717516 | 1.65 |
ENST00000200181.3
ENST00000339591.3 |
ITGB4
|
integrin, beta 4 |
| chr4_+_159727272 | 1.64 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr1_-_93257951 | 1.61 |
ENST00000543509.1
ENST00000370331.1 ENST00000540033.1 |
EVI5
|
ecotropic viral integration site 5 |
| chr3_+_11178779 | 1.60 |
ENST00000438284.2
|
HRH1
|
histamine receptor H1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOSL2_SMARCC1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:1904303 | positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
| 2.2 | 11.0 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 2.2 | 13.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 2.1 | 6.3 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 1.6 | 4.7 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 1.6 | 29.7 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 1.5 | 9.0 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 1.5 | 13.5 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 1.5 | 10.3 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 1.4 | 49.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 1.1 | 3.3 | GO:0035698 | CD8-positive, alpha-beta T cell extravasation(GO:0035697) CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) positive regulation of necroptotic process(GO:0060545) regulation of T cell extravasation(GO:2000407) regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 1.1 | 11.7 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.1 | 3.2 | GO:0048627 | myoblast development(GO:0048627) |
| 1.1 | 3.2 | GO:1902362 | melanocyte apoptotic process(GO:1902362) |
| 1.0 | 4.0 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 1.0 | 25.9 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.9 | 4.7 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.9 | 4.6 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.9 | 8.2 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.9 | 2.7 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 0.8 | 6.8 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.8 | 3.3 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.8 | 2.4 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.8 | 7.9 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.8 | 6.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.8 | 6.9 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.8 | 3.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.7 | 2.9 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.7 | 3.6 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) neuron projection maintenance(GO:1990535) |
| 0.7 | 3.9 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.6 | 3.8 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.6 | 6.4 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.6 | 4.5 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.6 | 1.9 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.6 | 7.9 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.6 | 8.9 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.6 | 3.5 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.6 | 12.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.6 | 11.4 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.6 | 8.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.5 | 2.7 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.5 | 2.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.5 | 4.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.5 | 14.2 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.5 | 1.5 | GO:0002661 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.5 | 1.5 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of osteoclast proliferation(GO:0090291) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.5 | 2.4 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.5 | 1.9 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.5 | 2.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.5 | 3.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.4 | 2.7 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.4 | 1.7 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.4 | 3.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.4 | 1.5 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.4 | 4.9 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.4 | 2.6 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.4 | 10.4 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.4 | 4.3 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.4 | 1.1 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.3 | 2.4 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.3 | 2.0 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.3 | 4.7 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.3 | 6.9 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.3 | 4.2 | GO:0007320 | insemination(GO:0007320) |
| 0.3 | 0.9 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.3 | 2.0 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.3 | 2.3 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.3 | 0.5 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.3 | 1.1 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.3 | 2.8 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.3 | 1.0 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.2 | 1.2 | GO:1902722 | positive regulation of prolactin secretion(GO:1902722) |
| 0.2 | 1.0 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.2 | 1.4 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.2 | 1.4 | GO:0030421 | defecation(GO:0030421) |
| 0.2 | 1.6 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.2 | 3.7 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 1.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 1.3 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.2 | 1.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.2 | 4.1 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 6.5 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 | 0.6 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.2 | 1.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.2 | 0.6 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.2 | 3.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 1.8 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 0.4 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.2 | 0.9 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 | 0.9 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.2 | 5.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 2.0 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.2 | 0.8 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.2 | 1.5 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.2 | 5.0 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.2 | 4.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 1.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 3.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 2.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 1.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 1.8 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.4 | GO:0019082 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.1 | 0.6 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 4.1 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.8 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.4 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 2.4 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 1.8 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 3.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 1.7 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.8 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 0.9 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.1 | 1.5 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.8 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.4 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 1.1 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.1 | 1.8 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.1 | 1.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.9 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.1 | 2.4 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 1.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 2.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.3 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 0.9 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 0.6 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 0.7 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.5 | GO:0061502 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.5 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 1.3 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 0.9 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.8 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.5 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.4 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 8.8 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) |
| 0.1 | 1.0 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 2.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.7 | GO:0032960 | regulation of inositol trisphosphate biosynthetic process(GO:0032960) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.1 | 0.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 1.6 | GO:0035588 | G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 0.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.1 | 1.7 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 1.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 5.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 2.1 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 0.6 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.1 | 1.3 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.1 | 0.2 | GO:0006043 | glucosamine catabolic process(GO:0006043) |
| 0.1 | 1.5 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.1 | 1.3 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 0.4 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.7 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.5 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 2.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 1.2 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 1.0 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.1 | 0.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.1 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 1.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.9 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 12.4 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.3 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 2.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 1.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.0 | 0.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.6 | GO:0040013 | negative regulation of locomotion(GO:0040013) |
| 0.0 | 2.4 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.5 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.9 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 0.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.6 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 1.0 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.6 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 3.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.3 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.2 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 1.0 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.3 | GO:0051957 | positive regulation of amino acid transport(GO:0051957) |
| 0.0 | 0.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 3.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.8 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.2 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.9 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.0 | 1.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.5 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 1.5 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 2.1 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.1 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.0 | 0.3 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.2 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 3.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.5 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.8 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.1 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.6 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 3.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) Schwann cell proliferation(GO:0014010) |
| 0.0 | 0.6 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.6 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 1.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.2 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.5 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.9 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 3.5 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.2 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.9 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.1 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.0 | 0.7 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 1.1 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.1 | GO:0038112 | interleukin-8-mediated signaling pathway(GO:0038112) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 2.5 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.8 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.4 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 1.5 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 1.9 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.3 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 1.2 | GO:0044819 | mitotic G1 DNA damage checkpoint(GO:0031571) mitotic G1/S transition checkpoint(GO:0044819) |
| 0.0 | 0.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.4 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 4.2 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 1.5 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.7 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 1.1 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 38.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 3.1 | 9.2 | GO:0097444 | spine apparatus(GO:0097444) |
| 2.1 | 6.3 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 1.2 | 4.9 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 1.0 | 2.9 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.8 | 14.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.8 | 2.3 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.7 | 3.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.6 | 3.8 | GO:0031673 | H zone(GO:0031673) |
| 0.6 | 3.7 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.6 | 2.8 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.5 | 1.5 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.5 | 1.8 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.4 | 4.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.3 | 2.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.3 | 3.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 8.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.3 | 21.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.3 | 5.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 2.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.2 | 1.2 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.2 | 4.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 7.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 0.9 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.2 | 1.7 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.2 | 1.0 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.2 | 4.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 4.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 0.5 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.1 | 1.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 8.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 14.2 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 5.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.5 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 5.0 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 0.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 2.0 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 1.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 1.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 1.8 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 2.8 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 4.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 1.8 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 0.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 2.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.9 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 0.7 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 5.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 1.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 8.2 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 3.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 5.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 13.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 3.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.4 | GO:1990462 | omegasome(GO:1990462) |
| 0.1 | 1.4 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 0.4 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 0.5 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.4 | GO:0036019 | endolysosome(GO:0036019) |
| 0.0 | 1.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 6.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 3.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 7.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 1.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 1.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 9.2 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 3.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 5.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 2.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 2.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.4 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 13.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 7.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.1 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.3 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 4.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.1 | GO:0098858 | actin-based cell projection(GO:0098858) |
| 0.0 | 1.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.4 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 1.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 1.7 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 3.9 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0002080 | acrosomal membrane(GO:0002080) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.3 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 2.0 | 14.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 1.6 | 4.7 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 1.3 | 5.3 | GO:1990175 | EH domain binding(GO:1990175) |
| 1.2 | 8.3 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 1.0 | 4.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 1.0 | 13.8 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.9 | 6.3 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.8 | 2.4 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.7 | 2.7 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.6 | 3.6 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.6 | 2.9 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.6 | 6.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.6 | 8.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.5 | 3.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.5 | 1.9 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.4 | 5.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.4 | 9.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.4 | 5.8 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.4 | 2.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.4 | 1.5 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.4 | 1.5 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.4 | 13.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 1.7 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.3 | 1.0 | GO:0086040 | sodium:proton antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086040) |
| 0.3 | 3.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.3 | 3.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 6.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.3 | 6.9 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.3 | 1.2 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.3 | 0.9 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.3 | 0.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.3 | 1.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.3 | 44.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 4.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 5.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.3 | 1.9 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.3 | 4.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.3 | 2.8 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.3 | 1.5 | GO:0070026 | nitric oxide binding(GO:0070026) |
| 0.2 | 3.9 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.2 | 1.2 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.2 | 1.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.2 | 3.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 1.6 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.2 | 1.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 1.8 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.2 | 3.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 0.8 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.2 | 7.1 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.2 | 4.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 4.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 0.5 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.2 | 3.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 1.0 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 1.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 1.8 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.2 | 0.5 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.2 | 0.9 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 3.0 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 4.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.6 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.7 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 11.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 18.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 5.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.6 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.1 | 1.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 1.5 | GO:0008526 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 5.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.7 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.5 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 2.4 | GO:0001164 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 1.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.6 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.1 | 1.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 1.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.3 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.1 | 1.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 3.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 4.9 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 22.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 3.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 18.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 1.0 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 1.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.2 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.1 | 0.4 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 1.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 3.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.8 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.3 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.2 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.1 | 9.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 0.5 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 1.5 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 2.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 1.1 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 1.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.4 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.6 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 1.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.9 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 1.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.0 | 1.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 3.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 1.0 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 1.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 1.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 2.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.7 | GO:0004175 | endopeptidase activity(GO:0004175) |
| 0.0 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 5.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 2.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 receptor activity(GO:0004918) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.8 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 1.7 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.3 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.4 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.7 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 1.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.0 | GO:0003676 | nucleic acid binding(GO:0003676) |
| 0.0 | 1.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 1.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.6 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 1.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 3.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.5 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 1.1 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 6.6 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.4 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 3.2 | GO:0003924 | GTPase activity(GO:0003924) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 48.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 25.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.2 | 4.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 13.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.2 | 1.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 15.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 4.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 7.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 6.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 6.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 3.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.8 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 8.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 4.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 3.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 4.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 4.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 1.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 8.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 2.0 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 3.4 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 6.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 2.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 6.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 13.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.4 | 8.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.4 | 6.3 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.3 | 57.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.3 | 7.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 18.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.2 | 4.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 3.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 8.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 5.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 4.0 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 3.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 3.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 3.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 7.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 2.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 4.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 1.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 3.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 7.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 2.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.3 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 2.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 2.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 2.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 4.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 3.7 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 1.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 2.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.9 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.8 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 1.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 3.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 1.3 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |