Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
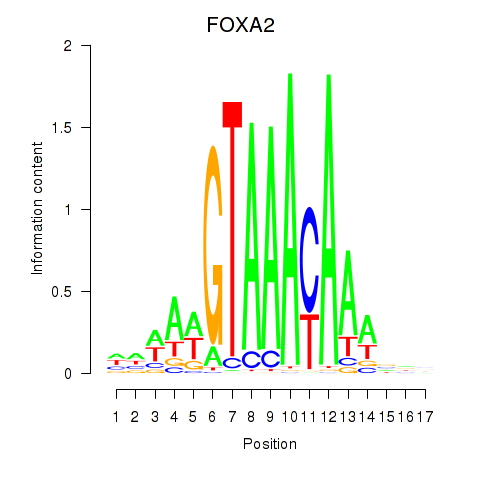
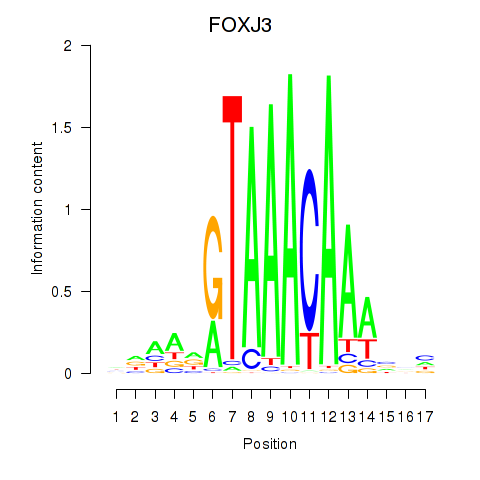
Results for FOXA2_FOXJ3
Z-value: 0.42
Transcription factors associated with FOXA2_FOXJ3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA2
|
ENSG00000125798.10 | forkhead box A2 |
|
FOXJ3
|
ENSG00000198815.4 | forkhead box J3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXA2 | hg19_v2_chr20_-_22565101_22565223 | 0.10 | 6.6e-01 | Click! |
| FOXJ3 | hg19_v2_chr1_-_42800860_42800912 | 0.09 | 7.2e-01 | Click! |
Activity profile of FOXA2_FOXJ3 motif
Sorted Z-values of FOXA2_FOXJ3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_137674000 | 0.56 |
ENST00000510119.1
ENST00000513970.1 |
CDC25C
|
cell division cycle 25C |
| chr5_+_169011033 | 0.55 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr16_+_11439286 | 0.55 |
ENST00000312499.5
ENST00000576027.1 |
RMI2
|
RecQ mediated genome instability 2 |
| chr12_-_106480587 | 0.53 |
ENST00000548902.1
|
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr10_+_70847852 | 0.49 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr3_-_58523010 | 0.44 |
ENST00000459701.2
ENST00000302819.5 |
ACOX2
|
acyl-CoA oxidase 2, branched chain |
| chr1_+_203765437 | 0.44 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr2_+_109223595 | 0.41 |
ENST00000410093.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr4_+_103790462 | 0.41 |
ENST00000503643.1
|
CISD2
|
CDGSH iron sulfur domain 2 |
| chr6_-_152623231 | 0.40 |
ENST00000540663.1
ENST00000537033.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr1_+_66796401 | 0.38 |
ENST00000528771.1
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr12_-_102591604 | 0.38 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr8_-_80993010 | 0.37 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr15_+_57998923 | 0.37 |
ENST00000380557.4
|
POLR2M
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr8_+_31497271 | 0.36 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chrX_+_9431324 | 0.35 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr12_-_76462713 | 0.34 |
ENST00000552056.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr1_-_238108575 | 0.33 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr17_+_71229346 | 0.33 |
ENST00000535032.2
ENST00000582793.1 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr2_+_74120094 | 0.31 |
ENST00000409731.3
ENST00000345517.3 ENST00000409918.1 ENST00000442912.1 ENST00000409624.1 |
ACTG2
|
actin, gamma 2, smooth muscle, enteric |
| chr15_+_78857870 | 0.30 |
ENST00000559554.1
|
CHRNA5
|
cholinergic receptor, nicotinic, alpha 5 (neuronal) |
| chr12_+_20968608 | 0.30 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr9_-_86432547 | 0.30 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr2_+_109204909 | 0.28 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr8_-_40755333 | 0.28 |
ENST00000297737.6
ENST00000315769.7 |
ZMAT4
|
zinc finger, matrin-type 4 |
| chr4_-_174256276 | 0.28 |
ENST00000296503.5
|
HMGB2
|
high mobility group box 2 |
| chr3_+_172468749 | 0.27 |
ENST00000366254.2
ENST00000415665.1 ENST00000438041.1 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr6_-_131321863 | 0.26 |
ENST00000528282.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr15_+_78857849 | 0.26 |
ENST00000299565.5
|
CHRNA5
|
cholinergic receptor, nicotinic, alpha 5 (neuronal) |
| chr11_-_85780853 | 0.26 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr12_-_12837423 | 0.25 |
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19 |
| chr17_-_41985096 | 0.24 |
ENST00000269095.4
ENST00000523220.1 |
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr16_+_53469525 | 0.24 |
ENST00000544405.2
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr4_-_111120334 | 0.23 |
ENST00000503885.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr15_+_69857515 | 0.23 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr11_+_65851443 | 0.23 |
ENST00000533756.1
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr17_-_30228678 | 0.23 |
ENST00000261708.4
|
UTP6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr7_+_26332645 | 0.23 |
ENST00000396376.1
|
SNX10
|
sorting nexin 10 |
| chr10_+_94352956 | 0.23 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr6_+_62284008 | 0.22 |
ENST00000544932.1
|
MTRNR2L9
|
MT-RNR2-like 9 (pseudogene) |
| chr9_-_4299874 | 0.22 |
ENST00000381971.3
ENST00000477901.1 |
GLIS3
|
GLIS family zinc finger 3 |
| chr1_+_95616933 | 0.22 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr8_+_27631903 | 0.22 |
ENST00000305188.8
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr9_-_94877658 | 0.21 |
ENST00000262554.2
ENST00000337841.4 |
SPTLC1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr1_-_246729544 | 0.21 |
ENST00000544618.1
ENST00000366514.4 |
TFB2M
|
transcription factor B2, mitochondrial |
| chr7_-_81635106 | 0.21 |
ENST00000443883.1
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr12_-_12714006 | 0.20 |
ENST00000541207.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr5_-_98262240 | 0.20 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr17_-_57229155 | 0.20 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr21_-_16374688 | 0.20 |
ENST00000411932.1
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr14_-_102605983 | 0.20 |
ENST00000334701.7
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chrX_-_47863348 | 0.20 |
ENST00000376943.3
ENST00000396965.1 ENST00000305127.6 |
ZNF182
|
zinc finger protein 182 |
| chr5_-_159846066 | 0.20 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr16_+_10479906 | 0.19 |
ENST00000562527.1
ENST00000396560.2 ENST00000396559.1 ENST00000562102.1 ENST00000543967.1 ENST00000569939.1 ENST00000569900.1 |
ATF7IP2
|
activating transcription factor 7 interacting protein 2 |
| chr7_+_112063192 | 0.19 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr14_+_97263641 | 0.19 |
ENST00000216639.3
|
VRK1
|
vaccinia related kinase 1 |
| chr5_+_159436120 | 0.19 |
ENST00000522793.1
ENST00000231238.5 |
TTC1
|
tetratricopeptide repeat domain 1 |
| chr19_-_10311868 | 0.19 |
ENST00000588118.1
ENST00000586800.1 |
DNMT1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr10_-_32217717 | 0.19 |
ENST00000396144.4
ENST00000375245.4 ENST00000344936.2 ENST00000375250.5 |
ARHGAP12
|
Rho GTPase activating protein 12 |
| chr6_-_13621126 | 0.19 |
ENST00000600057.1
|
AL441883.1
|
Uncharacterized protein |
| chr5_+_65222500 | 0.18 |
ENST00000511297.1
ENST00000506030.1 |
ERBB2IP
|
erbb2 interacting protein |
| chr1_+_161035655 | 0.18 |
ENST00000600454.1
|
AL591806.1
|
Uncharacterized protein |
| chr3_-_185641681 | 0.18 |
ENST00000259043.7
|
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chr14_+_32547434 | 0.18 |
ENST00000556191.1
ENST00000554090.1 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr17_-_26662464 | 0.18 |
ENST00000579419.1
ENST00000585313.1 ENST00000395418.3 ENST00000578985.1 ENST00000577498.1 ENST00000585089.1 ENST00000357896.3 |
IFT20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr14_+_74551650 | 0.18 |
ENST00000554938.1
|
LIN52
|
lin-52 homolog (C. elegans) |
| chr3_+_69928256 | 0.18 |
ENST00000394355.2
|
MITF
|
microphthalmia-associated transcription factor |
| chr9_-_39288092 | 0.18 |
ENST00000323947.7
ENST00000297668.6 ENST00000377656.2 ENST00000377659.1 |
CNTNAP3
|
contactin associated protein-like 3 |
| chr5_+_179135246 | 0.18 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr2_+_46844290 | 0.18 |
ENST00000238892.3
|
CRIPT
|
cysteine-rich PDZ-binding protein |
| chr2_-_128615681 | 0.17 |
ENST00000409955.1
ENST00000272645.4 |
POLR2D
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr4_-_170679024 | 0.17 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
| chr12_+_27175476 | 0.17 |
ENST00000546323.1
ENST00000282892.3 |
MED21
|
mediator complex subunit 21 |
| chr10_+_62538089 | 0.17 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr3_+_164924716 | 0.17 |
ENST00000470138.1
ENST00000498616.1 |
RP11-85M11.2
|
RP11-85M11.2 |
| chr15_-_54051831 | 0.17 |
ENST00000557913.1
ENST00000360509.5 |
WDR72
|
WD repeat domain 72 |
| chr8_+_27632047 | 0.17 |
ENST00000397418.2
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr8_+_27632083 | 0.16 |
ENST00000519637.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr2_+_109204743 | 0.16 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr12_+_72080253 | 0.16 |
ENST00000549735.1
|
TMEM19
|
transmembrane protein 19 |
| chr7_-_30009542 | 0.16 |
ENST00000438497.1
|
SCRN1
|
secernin 1 |
| chr3_+_142315294 | 0.16 |
ENST00000464320.1
|
PLS1
|
plastin 1 |
| chr12_+_79439405 | 0.16 |
ENST00000552744.1
|
SYT1
|
synaptotagmin I |
| chr11_-_92930556 | 0.16 |
ENST00000529184.1
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr3_-_182703688 | 0.16 |
ENST00000466812.1
ENST00000487822.1 ENST00000460412.1 ENST00000469954.1 |
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr2_-_37193606 | 0.16 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chr17_-_1418972 | 0.15 |
ENST00000571274.1
|
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr9_-_21305312 | 0.15 |
ENST00000259555.4
|
IFNA5
|
interferon, alpha 5 |
| chr8_-_101321584 | 0.15 |
ENST00000523167.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr3_+_142315225 | 0.15 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chr2_-_128615517 | 0.15 |
ENST00000409698.1
|
POLR2D
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr19_+_41768561 | 0.15 |
ENST00000599719.1
ENST00000601309.1 |
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr1_+_246729815 | 0.15 |
ENST00000366511.1
|
CNST
|
consortin, connexin sorting protein |
| chr12_+_49297899 | 0.14 |
ENST00000552942.1
ENST00000320516.4 |
CCDC65
|
coiled-coil domain containing 65 |
| chr9_-_5830768 | 0.14 |
ENST00000381506.3
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr15_+_35270552 | 0.14 |
ENST00000391457.2
|
AC114546.1
|
HCG37415; PRO1914; Uncharacterized protein |
| chr2_+_162165038 | 0.14 |
ENST00000437630.1
|
PSMD14
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 |
| chr7_-_16840820 | 0.14 |
ENST00000450569.1
|
AGR2
|
anterior gradient 2 |
| chr2_-_26251481 | 0.14 |
ENST00000599234.1
|
AC013449.1
|
Uncharacterized protein |
| chr16_-_80926457 | 0.14 |
ENST00000563626.1
ENST00000562231.1 |
RP11-314O13.1
|
RP11-314O13.1 |
| chr5_-_59064458 | 0.14 |
ENST00000502575.1
ENST00000507116.1 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr5_+_136070614 | 0.14 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr8_+_21777159 | 0.14 |
ENST00000434536.1
ENST00000252512.9 |
XPO7
|
exportin 7 |
| chr2_+_65283506 | 0.13 |
ENST00000377990.2
|
CEP68
|
centrosomal protein 68kDa |
| chr2_+_149402989 | 0.13 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr7_-_30008849 | 0.13 |
ENST00000409497.1
|
SCRN1
|
secernin 1 |
| chr16_+_22517166 | 0.13 |
ENST00000356156.3
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr2_+_233527443 | 0.13 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr5_-_58571935 | 0.13 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr5_-_10308125 | 0.13 |
ENST00000296658.3
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr14_-_75518129 | 0.13 |
ENST00000556257.1
ENST00000557648.1 ENST00000553263.1 ENST00000355774.2 ENST00000380968.2 ENST00000238662.7 |
MLH3
|
mutL homolog 3 |
| chr6_+_4087664 | 0.13 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chr10_-_69597915 | 0.13 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr9_+_108463234 | 0.13 |
ENST00000374688.1
|
TMEM38B
|
transmembrane protein 38B |
| chr12_+_54378923 | 0.13 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr9_-_136933615 | 0.13 |
ENST00000371834.2
|
BRD3
|
bromodomain containing 3 |
| chr15_+_99433570 | 0.13 |
ENST00000558898.1
|
IGF1R
|
insulin-like growth factor 1 receptor |
| chr1_+_92545862 | 0.13 |
ENST00000370382.3
ENST00000342818.3 |
BTBD8
|
BTB (POZ) domain containing 8 |
| chr16_-_88772670 | 0.13 |
ENST00000562544.1
|
RNF166
|
ring finger protein 166 |
| chr7_+_151038850 | 0.13 |
ENST00000355851.4
ENST00000566856.1 ENST00000470229.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr9_-_20382446 | 0.13 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr7_+_6617039 | 0.12 |
ENST00000405731.3
ENST00000396713.2 ENST00000396707.2 ENST00000335965.6 ENST00000396709.1 ENST00000483589.1 ENST00000396706.2 |
ZDHHC4
|
zinc finger, DHHC-type containing 4 |
| chr9_-_119162885 | 0.12 |
ENST00000445861.2
|
PAPPA-AS1
|
PAPPA antisense RNA 1 |
| chr10_-_14050522 | 0.12 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr10_-_25010795 | 0.12 |
ENST00000416305.1
ENST00000376410.2 |
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr3_+_101292939 | 0.12 |
ENST00000265260.3
ENST00000469941.1 ENST00000296024.5 |
PCNP
|
PEST proteolytic signal containing nuclear protein |
| chr1_+_78470530 | 0.12 |
ENST00000370763.5
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr3_-_135916073 | 0.12 |
ENST00000481989.1
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chr19_-_40030861 | 0.12 |
ENST00000390658.2
|
EID2
|
EP300 interacting inhibitor of differentiation 2 |
| chr20_+_9049303 | 0.12 |
ENST00000407043.2
ENST00000441846.1 |
PLCB4
|
phospholipase C, beta 4 |
| chr12_-_15374343 | 0.12 |
ENST00000256953.2
ENST00000546331.1 |
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chr5_+_79703823 | 0.12 |
ENST00000338008.5
ENST00000510158.1 ENST00000505560.1 |
ZFYVE16
|
zinc finger, FYVE domain containing 16 |
| chr5_+_140186647 | 0.12 |
ENST00000512229.2
ENST00000356878.4 ENST00000530339.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr7_-_6098770 | 0.12 |
ENST00000536084.1
ENST00000446699.1 ENST00000199389.6 |
EIF2AK1
|
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr1_-_77685084 | 0.12 |
ENST00000370812.3
ENST00000359130.1 ENST00000445065.1 ENST00000370813.5 |
PIGK
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr14_-_58893832 | 0.12 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr3_+_148447887 | 0.12 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr17_-_26662440 | 0.12 |
ENST00000578122.1
|
IFT20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr2_-_113191096 | 0.12 |
ENST00000496537.1
ENST00000330575.5 ENST00000302558.3 |
RGPD8
|
RANBP2-like and GRIP domain containing 8 |
| chr10_-_69597828 | 0.12 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr12_+_97306295 | 0.12 |
ENST00000457368.2
|
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr7_+_77469439 | 0.12 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr1_+_203830703 | 0.11 |
ENST00000414487.2
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E |
| chr15_-_31283798 | 0.11 |
ENST00000435680.1
ENST00000425768.1 |
MTMR10
|
myotubularin related protein 10 |
| chr7_+_106809406 | 0.11 |
ENST00000468410.1
ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1
|
HMG-box transcription factor 1 |
| chr5_+_153418466 | 0.11 |
ENST00000522782.1
ENST00000439768.2 ENST00000436816.1 ENST00000322602.5 ENST00000522177.1 ENST00000520899.1 |
MFAP3
|
microfibrillar-associated protein 3 |
| chr14_+_35747825 | 0.11 |
ENST00000540871.1
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr6_-_42185583 | 0.11 |
ENST00000053468.3
|
MRPS10
|
mitochondrial ribosomal protein S10 |
| chr12_+_2986359 | 0.11 |
ENST00000538636.1
ENST00000461997.2 ENST00000489288.2 ENST00000366285.2 ENST00000538700.1 |
RHNO1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
| chr14_+_58894103 | 0.11 |
ENST00000354386.6
ENST00000556134.1 |
KIAA0586
|
KIAA0586 |
| chr9_+_80850952 | 0.11 |
ENST00000424347.2
ENST00000415759.2 ENST00000376597.4 ENST00000277082.5 ENST00000376598.2 |
CEP78
|
centrosomal protein 78kDa |
| chr1_+_84609944 | 0.11 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr13_+_41885341 | 0.11 |
ENST00000379406.3
ENST00000379367.3 ENST00000403412.3 |
NAA16
|
N(alpha)-acetyltransferase 16, NatA auxiliary subunit |
| chr11_+_102217936 | 0.11 |
ENST00000532832.1
ENST00000530675.1 ENST00000533742.1 ENST00000227758.2 ENST00000532672.1 ENST00000531259.1 ENST00000527465.1 |
BIRC2
|
baculoviral IAP repeat containing 2 |
| chr16_+_66442411 | 0.11 |
ENST00000499966.1
|
LINC00920
|
long intergenic non-protein coding RNA 920 |
| chr14_+_56078695 | 0.11 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr3_+_156393349 | 0.11 |
ENST00000473702.1
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr17_+_22022437 | 0.11 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr10_-_69597810 | 0.11 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr6_-_131291572 | 0.11 |
ENST00000529208.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr5_-_146833222 | 0.11 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr5_+_147774275 | 0.11 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr10_+_124739964 | 0.10 |
ENST00000406217.2
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr14_-_58894223 | 0.10 |
ENST00000555593.1
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr1_+_18958008 | 0.10 |
ENST00000420770.2
ENST00000400661.3 |
PAX7
|
paired box 7 |
| chr1_+_224370873 | 0.10 |
ENST00000323699.4
ENST00000391877.3 |
DEGS1
|
delta(4)-desaturase, sphingolipid 1 |
| chr2_+_143635067 | 0.10 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr18_+_21033239 | 0.10 |
ENST00000581585.1
ENST00000577501.1 |
RIOK3
|
RIO kinase 3 |
| chr14_-_58894332 | 0.10 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr15_-_59041768 | 0.10 |
ENST00000402627.1
ENST00000396140.2 ENST00000559053.1 ENST00000561288.1 |
ADAM10
|
ADAM metallopeptidase domain 10 |
| chr1_-_92371839 | 0.10 |
ENST00000370399.2
|
TGFBR3
|
transforming growth factor, beta receptor III |
| chr14_+_58894141 | 0.10 |
ENST00000423743.3
|
KIAA0586
|
KIAA0586 |
| chr1_-_114429997 | 0.10 |
ENST00000471267.1
ENST00000393320.3 |
BCL2L15
|
BCL2-like 15 |
| chr8_-_93978333 | 0.10 |
ENST00000524037.1
ENST00000520430.1 ENST00000521617.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr2_-_46844242 | 0.10 |
ENST00000281382.6
|
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr4_-_164534657 | 0.10 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr17_-_3595181 | 0.10 |
ENST00000552050.1
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr2_+_143635222 | 0.10 |
ENST00000375773.2
ENST00000409512.1 ENST00000410015.2 |
KYNU
|
kynureninase |
| chrX_+_120181457 | 0.10 |
ENST00000328078.1
|
GLUD2
|
glutamate dehydrogenase 2 |
| chr5_-_57854070 | 0.10 |
ENST00000504333.1
|
CTD-2117L12.1
|
Uncharacterized protein |
| chr19_-_52307357 | 0.10 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr14_-_22938665 | 0.10 |
ENST00000535880.2
|
TRDV3
|
T cell receptor delta variable 3 |
| chr5_-_60240858 | 0.09 |
ENST00000426742.2
ENST00000265038.5 ENST00000543101.1 ENST00000439176.1 |
ERCC8
|
excision repair cross-complementing rodent repair deficiency, complementation group 8 |
| chr17_+_67410832 | 0.09 |
ENST00000590474.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr2_+_172309634 | 0.09 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr5_+_71616188 | 0.09 |
ENST00000380639.5
ENST00000543322.1 ENST00000503868.1 ENST00000510676.2 ENST00000536805.1 |
PTCD2
|
pentatricopeptide repeat domain 2 |
| chr6_+_10528560 | 0.09 |
ENST00000379597.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr3_+_159570722 | 0.09 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr2_-_65593784 | 0.09 |
ENST00000443619.2
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr2_+_175260451 | 0.09 |
ENST00000458563.1
ENST00000409673.3 ENST00000272732.6 ENST00000435964.1 |
SCRN3
|
secernin 3 |
| chr8_+_103540983 | 0.09 |
ENST00000523572.1
|
KB-1980E6.3
|
Uncharacterized protein |
| chr2_-_3521518 | 0.09 |
ENST00000382093.5
|
ADI1
|
acireductone dioxygenase 1 |
| chr2_-_240230890 | 0.09 |
ENST00000446876.1
|
HDAC4
|
histone deacetylase 4 |
| chr17_-_49021974 | 0.09 |
ENST00000501718.2
|
RP11-700H6.1
|
RP11-700H6.1 |
| chr12_+_32832203 | 0.09 |
ENST00000553257.1
ENST00000549701.1 ENST00000358214.5 ENST00000266481.6 ENST00000551476.1 ENST00000550154.1 ENST00000547312.1 ENST00000414834.2 ENST00000381000.4 ENST00000548750.1 |
DNM1L
|
dynamin 1-like |
| chr10_+_90672113 | 0.09 |
ENST00000371922.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr8_-_93978309 | 0.09 |
ENST00000517858.1
ENST00000378861.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr16_+_57847684 | 0.09 |
ENST00000335616.2
|
CTD-2600O9.1
|
uncharacterized protein LOC388282 |
| chr1_-_241799232 | 0.09 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2) |
| chr21_+_34602377 | 0.09 |
ENST00000342101.3
ENST00000413881.1 ENST00000443073.1 |
IFNAR2
|
interferon (alpha, beta and omega) receptor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA2_FOXJ3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.4 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.1 | 0.3 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.1 | 0.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.3 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.1 | 0.2 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.2 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.2 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.2 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.6 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.4 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.5 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.2 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.6 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.3 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.0 | 0.2 | GO:0034505 | tooth mineralization(GO:0034505) enamel mineralization(GO:0070166) |
| 0.0 | 0.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.4 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.3 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.6 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.4 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.1 | GO:0060621 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.0 | 0.1 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.1 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.2 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.2 | GO:2001151 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.0 | 0.1 | GO:2000627 | rRNA import into mitochondrion(GO:0035928) regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.0 | 0.2 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.2 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.3 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.4 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0090309 | maintenance of DNA methylation(GO:0010216) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.5 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:0030806 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.4 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.0 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:1902510 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.8 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.0 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.4 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.5 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.5 | GO:0000732 | strand displacement(GO:0000732) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.1 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:1902741 | interferon-alpha secretion(GO:0072642) telomerase RNA stabilization(GO:0090669) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 0.1 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.2 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.4 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.3 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.0 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.1 | 0.2 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.1 | 0.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.3 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.3 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.1 | 0.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.2 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.1 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.0 | 0.2 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.4 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.0 | 0.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.9 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.0 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |