Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
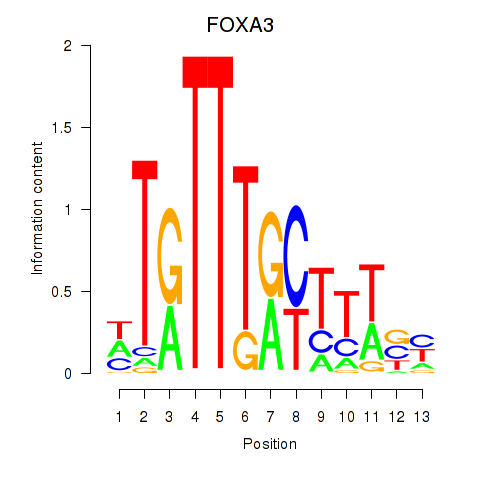
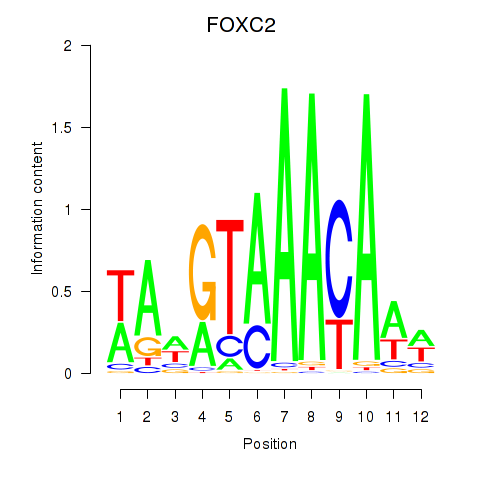
Results for FOXA3_FOXC2
Z-value: 0.67
Transcription factors associated with FOXA3_FOXC2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA3
|
ENSG00000170608.2 | forkhead box A3 |
|
FOXC2
|
ENSG00000176692.4 | forkhead box C2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXA3 | hg19_v2_chr19_+_46367518_46367567 | 0.77 | 7.3e-05 | Click! |
| FOXC2 | hg19_v2_chr16_+_86600857_86600921 | 0.69 | 7.5e-04 | Click! |
Activity profile of FOXA3_FOXC2 motif
Sorted Z-values of FOXA3_FOXC2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_25616519 | 2.66 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr9_-_75567962 | 2.42 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr5_-_137674000 | 1.77 |
ENST00000510119.1
ENST00000513970.1 |
CDC25C
|
cell division cycle 25C |
| chr17_+_79953310 | 1.75 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr2_+_128175997 | 1.71 |
ENST00000234071.3
ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr10_+_70847852 | 1.71 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr17_+_72426891 | 1.61 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr17_-_26695013 | 1.55 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr7_+_134832808 | 1.54 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr2_+_74120094 | 1.51 |
ENST00000409731.3
ENST00000345517.3 ENST00000409918.1 ENST00000442912.1 ENST00000409624.1 |
ACTG2
|
actin, gamma 2, smooth muscle, enteric |
| chr11_-_102401469 | 1.45 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr2_-_165424973 | 1.44 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr4_-_174256276 | 1.43 |
ENST00000296503.5
|
HMGB2
|
high mobility group box 2 |
| chrX_+_9431324 | 1.37 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr3_-_186080012 | 1.35 |
ENST00000544847.1
ENST00000265022.3 |
DGKG
|
diacylglycerol kinase, gamma 90kDa |
| chr17_-_26694979 | 1.35 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr2_-_175711133 | 1.32 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr17_+_4613918 | 1.27 |
ENST00000574954.1
ENST00000346341.2 ENST00000572457.1 ENST00000381488.6 ENST00000412477.3 ENST00000571428.1 ENST00000575877.1 |
ARRB2
|
arrestin, beta 2 |
| chr10_-_118032979 | 1.22 |
ENST00000355422.6
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr21_-_43786634 | 1.17 |
ENST00000291527.2
|
TFF1
|
trefoil factor 1 |
| chr10_-_118032697 | 1.12 |
ENST00000439649.3
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr11_+_8040739 | 1.06 |
ENST00000534099.1
|
TUB
|
tubby bipartite transcription factor |
| chr5_+_156712372 | 1.06 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr17_-_26697304 | 1.06 |
ENST00000536498.1
|
VTN
|
vitronectin |
| chr11_+_65657875 | 1.05 |
ENST00000312579.2
|
CCDC85B
|
coiled-coil domain containing 85B |
| chr17_+_4613776 | 1.05 |
ENST00000269260.2
|
ARRB2
|
arrestin, beta 2 |
| chr9_-_4299874 | 1.03 |
ENST00000381971.3
ENST00000477901.1 |
GLIS3
|
GLIS family zinc finger 3 |
| chr5_+_169011033 | 1.00 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr11_-_115375107 | 1.00 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr7_+_106415457 | 0.92 |
ENST00000490162.2
ENST00000470135.1 |
RP5-884M6.1
|
RP5-884M6.1 |
| chr2_+_46844290 | 0.91 |
ENST00000238892.3
|
CRIPT
|
cysteine-rich PDZ-binding protein |
| chr19_+_782755 | 0.91 |
ENST00000606242.1
ENST00000586061.1 |
AC006273.5
|
AC006273.5 |
| chr4_-_164534657 | 0.89 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr17_-_57229155 | 0.89 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr11_+_34999328 | 0.88 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr17_-_46688334 | 0.88 |
ENST00000239165.7
|
HOXB7
|
homeobox B7 |
| chr14_-_21562648 | 0.86 |
ENST00000555270.1
|
ZNF219
|
zinc finger protein 219 |
| chr5_+_133859996 | 0.86 |
ENST00000512386.1
|
PHF15
|
jade family PHD finger 2 |
| chr14_+_21156915 | 0.85 |
ENST00000397990.4
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr7_-_107968921 | 0.84 |
ENST00000442580.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr5_-_58571935 | 0.84 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr17_-_202579 | 0.82 |
ENST00000577079.1
ENST00000331302.7 ENST00000536489.2 |
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr1_+_200011711 | 0.82 |
ENST00000544748.1
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr8_-_145754428 | 0.75 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr14_-_94789663 | 0.73 |
ENST00000557225.1
ENST00000341584.3 |
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr19_+_54368001 | 0.72 |
ENST00000422045.1
|
AC008440.10
|
AC008440.10 |
| chr10_+_95848824 | 0.71 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr1_-_149459549 | 0.71 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C |
| chr1_-_246729544 | 0.71 |
ENST00000544618.1
ENST00000366514.4 |
TFB2M
|
transcription factor B2, mitochondrial |
| chr17_-_26733604 | 0.71 |
ENST00000584426.1
ENST00000584995.1 |
SLC46A1
|
solute carrier family 46 (folate transporter), member 1 |
| chr15_+_69857515 | 0.70 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chrX_-_15402498 | 0.70 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr2_-_192711968 | 0.69 |
ENST00000304141.4
|
SDPR
|
serum deprivation response |
| chr7_+_95115210 | 0.66 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr12_-_102591604 | 0.65 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr19_+_42806250 | 0.63 |
ENST00000598490.1
ENST00000341747.3 |
PRR19
|
proline rich 19 |
| chr15_+_59910132 | 0.63 |
ENST00000559200.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr1_+_66820058 | 0.63 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr16_+_77233294 | 0.63 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr11_-_107582775 | 0.61 |
ENST00000305991.2
|
SLN
|
sarcolipin |
| chr18_+_3449330 | 0.61 |
ENST00000549253.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr16_-_67597789 | 0.59 |
ENST00000605277.1
|
CTD-2012K14.6
|
CTD-2012K14.6 |
| chr17_+_72427477 | 0.59 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr16_+_84801852 | 0.58 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr1_+_18958008 | 0.58 |
ENST00000420770.2
ENST00000400661.3 |
PAX7
|
paired box 7 |
| chr16_+_20775358 | 0.58 |
ENST00000440284.2
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr17_-_26733179 | 0.57 |
ENST00000440501.1
ENST00000321666.5 |
SLC46A1
|
solute carrier family 46 (folate transporter), member 1 |
| chr14_+_23299088 | 0.57 |
ENST00000355151.5
ENST00000397496.3 ENST00000555345.1 ENST00000432849.3 ENST00000553711.1 ENST00000556465.1 ENST00000397505.2 ENST00000557221.1 ENST00000311892.6 ENST00000556840.1 ENST00000555536.1 |
MRPL52
|
mitochondrial ribosomal protein L52 |
| chr6_+_142623758 | 0.57 |
ENST00000541199.1
ENST00000435011.2 |
GPR126
|
G protein-coupled receptor 126 |
| chrX_-_20236970 | 0.56 |
ENST00000379548.4
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr2_-_145275228 | 0.54 |
ENST00000427902.1
ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr8_-_95449155 | 0.52 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr12_-_92539614 | 0.52 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr7_+_151791037 | 0.51 |
ENST00000419245.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr8_+_19171196 | 0.51 |
ENST00000518040.1
|
SH2D4A
|
SH2 domain containing 4A |
| chr10_-_46030841 | 0.50 |
ENST00000453424.2
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
| chr2_+_169658928 | 0.50 |
ENST00000317647.7
ENST00000445023.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr5_-_59481406 | 0.49 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr2_+_169659121 | 0.48 |
ENST00000397206.2
ENST00000397209.2 ENST00000421711.2 |
NOSTRIN
|
nitric oxide synthase trafficking |
| chr4_+_15376165 | 0.48 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr2_-_175711978 | 0.48 |
ENST00000409089.2
|
CHN1
|
chimerin 1 |
| chr17_-_39041479 | 0.47 |
ENST00000167588.3
|
KRT20
|
keratin 20 |
| chr15_+_59665194 | 0.47 |
ENST00000560394.1
|
FAM81A
|
family with sequence similarity 81, member A |
| chr6_-_165989936 | 0.46 |
ENST00000354448.4
|
PDE10A
|
phosphodiesterase 10A |
| chr2_-_46844159 | 0.46 |
ENST00000474980.1
ENST00000306465.4 |
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr1_+_244214577 | 0.46 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr7_+_77469439 | 0.46 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr6_-_42016385 | 0.45 |
ENST00000502771.1
ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3
|
cyclin D3 |
| chr18_+_68002675 | 0.45 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr17_-_79212825 | 0.45 |
ENST00000374769.2
|
ENTHD2
|
ENTH domain containing 2 |
| chr3_+_119316721 | 0.45 |
ENST00000488919.1
ENST00000495992.1 |
PLA1A
|
phospholipase A1 member A |
| chr3_+_148447887 | 0.45 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr14_+_38065052 | 0.45 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr15_+_59664884 | 0.44 |
ENST00000558348.1
|
FAM81A
|
family with sequence similarity 81, member A |
| chr7_+_112063192 | 0.44 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr5_+_142286887 | 0.43 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr4_-_103266626 | 0.43 |
ENST00000356736.4
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr10_+_35484053 | 0.43 |
ENST00000487763.1
ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM
|
cAMP responsive element modulator |
| chr17_-_19290483 | 0.42 |
ENST00000395592.2
ENST00000299610.4 |
MFAP4
|
microfibrillar-associated protein 4 |
| chr8_+_101349823 | 0.42 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr14_+_74034310 | 0.42 |
ENST00000538782.1
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr5_+_60933634 | 0.42 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr7_-_107443652 | 0.42 |
ENST00000340010.5
ENST00000422236.2 ENST00000453332.1 |
SLC26A3
|
solute carrier family 26 (anion exchanger), member 3 |
| chr3_+_119316689 | 0.42 |
ENST00000273371.4
|
PLA1A
|
phospholipase A1 member A |
| chr3_+_50712672 | 0.42 |
ENST00000266037.9
|
DOCK3
|
dedicator of cytokinesis 3 |
| chrX_-_20237059 | 0.41 |
ENST00000457145.1
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr17_-_79212884 | 0.41 |
ENST00000300714.3
|
ENTHD2
|
ENTH domain containing 2 |
| chr15_-_59041954 | 0.41 |
ENST00000439637.1
ENST00000558004.1 |
ADAM10
|
ADAM metallopeptidase domain 10 |
| chr14_-_23299009 | 0.40 |
ENST00000488800.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr14_-_31926623 | 0.39 |
ENST00000356180.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr7_-_81635106 | 0.39 |
ENST00000443883.1
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr12_-_15865844 | 0.39 |
ENST00000543612.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr7_+_151791074 | 0.39 |
ENST00000447796.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr14_+_56585048 | 0.38 |
ENST00000267460.4
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr4_-_170679024 | 0.37 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
| chr12_+_72080253 | 0.36 |
ENST00000549735.1
|
TMEM19
|
transmembrane protein 19 |
| chr9_+_67977438 | 0.36 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr10_-_74283694 | 0.36 |
ENST00000398763.4
ENST00000418483.2 ENST00000489666.2 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr2_+_196521845 | 0.36 |
ENST00000359634.5
ENST00000412905.1 |
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr10_-_31288398 | 0.36 |
ENST00000538351.2
|
ZNF438
|
zinc finger protein 438 |
| chr14_+_56584414 | 0.36 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr1_-_54405773 | 0.36 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr14_-_31926701 | 0.35 |
ENST00000310850.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr8_-_80993010 | 0.35 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr5_+_140529630 | 0.35 |
ENST00000543635.1
|
PCDHB6
|
protocadherin beta 6 |
| chr15_-_59665062 | 0.35 |
ENST00000288235.4
|
MYO1E
|
myosin IE |
| chr14_-_101036119 | 0.35 |
ENST00000355173.2
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr17_-_27038063 | 0.35 |
ENST00000439862.3
|
PROCA1
|
protein interacting with cyclin A1 |
| chr8_-_23261589 | 0.34 |
ENST00000524168.1
ENST00000523833.2 ENST00000519243.1 ENST00000389131.3 |
LOXL2
|
lysyl oxidase-like 2 |
| chr8_+_19171128 | 0.34 |
ENST00000265807.3
|
SH2D4A
|
SH2 domain containing 4A |
| chr14_+_56127989 | 0.34 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr14_+_58711539 | 0.34 |
ENST00000216455.4
ENST00000412908.2 ENST00000557508.1 |
PSMA3
|
proteasome (prosome, macropain) subunit, alpha type, 3 |
| chr2_+_196522032 | 0.34 |
ENST00000418005.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr2_-_46844242 | 0.34 |
ENST00000281382.6
|
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chrX_+_135730373 | 0.33 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr2_+_162165038 | 0.33 |
ENST00000437630.1
|
PSMD14
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 |
| chr15_+_80364901 | 0.33 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chrX_+_135730297 | 0.33 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr3_+_69985734 | 0.33 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr14_+_20215587 | 0.32 |
ENST00000331723.1
|
OR4Q3
|
olfactory receptor, family 4, subfamily Q, member 3 |
| chr19_+_50380917 | 0.32 |
ENST00000535102.2
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr5_+_61874562 | 0.32 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr4_+_76871883 | 0.32 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr2_-_145275828 | 0.32 |
ENST00000392861.2
ENST00000409211.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr12_-_106480587 | 0.31 |
ENST00000548902.1
|
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr3_+_141106643 | 0.31 |
ENST00000514251.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr19_+_50380682 | 0.30 |
ENST00000221543.5
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr3_+_148545586 | 0.30 |
ENST00000282957.4
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 (tissue) |
| chr6_+_10528560 | 0.30 |
ENST00000379597.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chrX_+_10124977 | 0.30 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr20_+_11898507 | 0.30 |
ENST00000378226.2
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr1_-_201096312 | 0.30 |
ENST00000449188.2
|
ASCL5
|
achaete-scute family bHLH transcription factor 5 |
| chr15_-_50411412 | 0.29 |
ENST00000284509.6
|
ATP8B4
|
ATPase, class I, type 8B, member 4 |
| chr3_-_45957534 | 0.29 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr12_+_94071129 | 0.29 |
ENST00000552983.1
ENST00000332896.3 ENST00000552033.1 ENST00000548483.1 |
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr8_+_104384616 | 0.29 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr3_+_158991025 | 0.29 |
ENST00000337808.6
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr17_+_79213039 | 0.29 |
ENST00000431388.2
|
C17orf89
|
chromosome 17 open reading frame 89 |
| chr9_-_28670283 | 0.28 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr4_+_57138437 | 0.28 |
ENST00000504228.1
ENST00000541073.1 |
KIAA1211
|
KIAA1211 |
| chr9_+_129097520 | 0.28 |
ENST00000436593.3
|
MVB12B
|
multivesicular body subunit 12B |
| chr3_-_45957088 | 0.27 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr12_+_94071341 | 0.27 |
ENST00000542893.2
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr17_-_77967433 | 0.27 |
ENST00000571872.1
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr13_-_36429763 | 0.27 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr2_-_86422523 | 0.27 |
ENST00000442664.2
ENST00000409051.2 ENST00000449247.2 |
IMMT
|
inner membrane protein, mitochondrial |
| chr3_+_142315225 | 0.26 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chr15_+_49462434 | 0.26 |
ENST00000558145.1
ENST00000543495.1 ENST00000544523.1 ENST00000560138.1 |
GALK2
|
galactokinase 2 |
| chr7_+_26438187 | 0.26 |
ENST00000439120.1
ENST00000430548.1 ENST00000421862.1 ENST00000449537.1 ENST00000420774.1 ENST00000418758.2 |
AC004540.5
|
AC004540.5 |
| chr12_-_57328187 | 0.26 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr15_+_75335604 | 0.25 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr19_-_51141196 | 0.25 |
ENST00000338916.4
|
SYT3
|
synaptotagmin III |
| chr8_-_30002179 | 0.25 |
ENST00000320542.3
|
MBOAT4
|
membrane bound O-acyltransferase domain containing 4 |
| chr14_-_23526739 | 0.25 |
ENST00000397359.3
ENST00000487137.2 |
CDH24
|
cadherin 24, type 2 |
| chr4_-_140223670 | 0.24 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr2_+_196521903 | 0.24 |
ENST00000541054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr1_-_167522982 | 0.24 |
ENST00000370509.4
|
CREG1
|
cellular repressor of E1A-stimulated genes 1 |
| chr3_+_137728842 | 0.24 |
ENST00000183605.5
|
CLDN18
|
claudin 18 |
| chr17_+_7487146 | 0.24 |
ENST00000396501.4
ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1
|
mannose-P-dolichol utilization defect 1 |
| chr5_+_161494770 | 0.24 |
ENST00000414552.2
ENST00000361925.4 |
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr7_-_107643567 | 0.24 |
ENST00000393559.1
ENST00000439976.1 ENST00000393560.1 |
LAMB1
|
laminin, beta 1 |
| chr14_+_74551650 | 0.24 |
ENST00000554938.1
|
LIN52
|
lin-52 homolog (C. elegans) |
| chr14_+_56127960 | 0.24 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr7_-_121944491 | 0.23 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr15_-_66797172 | 0.23 |
ENST00000569438.1
ENST00000569696.1 ENST00000307961.6 |
RPL4
|
ribosomal protein L4 |
| chr12_-_71551652 | 0.23 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr5_+_161495038 | 0.23 |
ENST00000393933.4
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr7_-_107643674 | 0.22 |
ENST00000222399.6
|
LAMB1
|
laminin, beta 1 |
| chr22_-_39268308 | 0.22 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr6_-_41715128 | 0.22 |
ENST00000356667.4
ENST00000373025.3 ENST00000425343.2 |
PGC
|
progastricsin (pepsinogen C) |
| chr2_+_136343904 | 0.22 |
ENST00000436436.1
|
R3HDM1
|
R3H domain containing 1 |
| chr7_+_106505696 | 0.21 |
ENST00000440650.2
ENST00000496166.1 ENST00000473541.1 |
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr1_-_43855479 | 0.21 |
ENST00000290663.6
ENST00000372457.4 |
MED8
|
mediator complex subunit 8 |
| chr15_+_71185148 | 0.21 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr15_+_69854027 | 0.21 |
ENST00000498938.2
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr1_+_207277590 | 0.21 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr7_+_106505912 | 0.21 |
ENST00000359195.3
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr1_+_84609944 | 0.21 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr12_-_122018346 | 0.21 |
ENST00000377069.4
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr3_-_107941230 | 0.21 |
ENST00000264538.3
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA3_FOXC2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.5 | 2.3 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.4 | 1.7 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.4 | 2.4 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.4 | 3.6 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.3 | 1.6 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.3 | 1.5 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 1.3 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.2 | 0.6 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.2 | 0.9 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.2 | 0.6 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.2 | 0.7 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 | 1.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 0.6 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.1 | 1.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 2.3 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.6 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.4 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 1.4 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 1.0 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.8 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.7 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 2.4 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.1 | 1.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 0.5 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 | 0.9 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 1.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.3 | GO:0018277 | protein deamination(GO:0018277) |
| 0.1 | 1.0 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 0.7 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 0.2 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.4 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.1 | 1.4 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.1 | 0.5 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.2 | GO:1903217 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.1 | 0.4 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.1 | 0.8 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.0 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.4 | GO:0038042 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) |
| 0.0 | 0.4 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.2 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.1 | GO:0039023 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.0 | GO:0021767 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) |
| 0.0 | 0.3 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.7 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.2 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.9 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 0.3 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.4 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 1.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.6 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.3 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.4 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.9 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 1.6 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.2 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.0 | 0.2 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.7 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 1.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 1.4 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.3 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.8 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.1 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.7 | GO:0051023 | regulation of immunoglobulin secretion(GO:0051023) |
| 0.0 | 0.7 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.0 | 0.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.5 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.6 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.7 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.1 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.2 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.4 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.5 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.5 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.2 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.0 | 1.7 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 1.0 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 1.0 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.0 | 0.7 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.3 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.6 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0046218 | tryptophan catabolic process(GO:0006569) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.2 | 0.5 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.1 | 0.9 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 2.7 | GO:0005916 | fascia adherens(GO:0005916) catenin complex(GO:0016342) |
| 0.1 | 0.4 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 1.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.9 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 1.9 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.7 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.3 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.1 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 1.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 2.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.3 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 2.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 2.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.6 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.0 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 2.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.3 | 1.4 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.3 | 2.4 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.3 | 2.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.2 | 0.7 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.2 | 0.7 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.2 | 1.3 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.2 | 0.6 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.2 | 0.8 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.4 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.4 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 0.4 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.1 | 0.9 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.1 | 0.9 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 0.7 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.9 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 2.7 | GO:0045294 | alpha-catenin binding(GO:0045294) gamma-catenin binding(GO:0045295) |
| 0.1 | 1.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 2.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.9 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 0.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.2 | GO:0070260 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.1 | 0.6 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 1.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.5 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.2 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 1.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.3 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 3.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.8 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 1.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.9 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 1.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 5.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.8 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 2.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 2.4 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 3.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 2.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.7 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 2.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.0 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 2.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.5 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.0 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |