Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
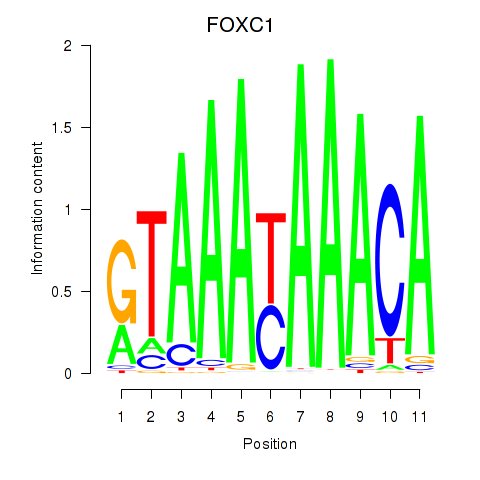
Results for FOXC1
Z-value: 0.62
Transcription factors associated with FOXC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXC1
|
ENSG00000054598.5 | forkhead box C1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXC1 | hg19_v2_chr6_+_1610681_1610681 | 0.46 | 4.4e-02 | Click! |
Activity profile of FOXC1 motif
Sorted Z-values of FOXC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_161056762 | 2.81 |
ENST00000428609.2
ENST00000409967.2 |
ITGB6
|
integrin, beta 6 |
| chr2_-_161056802 | 2.71 |
ENST00000283249.2
ENST00000409872.1 |
ITGB6
|
integrin, beta 6 |
| chr13_-_86373536 | 2.48 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr4_-_102267953 | 2.20 |
ENST00000523694.2
ENST00000507176.1 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr3_+_189349162 | 2.01 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr11_+_34654011 | 1.81 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr14_+_37126765 | 1.75 |
ENST00000402703.2
|
PAX9
|
paired box 9 |
| chr2_+_113885138 | 1.70 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr2_-_208030295 | 1.62 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr10_+_112631699 | 1.43 |
ENST00000444997.1
|
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor) |
| chr8_-_134309335 | 1.43 |
ENST00000522890.1
ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1
|
N-myc downstream regulated 1 |
| chr5_-_24645078 | 1.37 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr8_-_134309823 | 1.29 |
ENST00000414097.2
|
NDRG1
|
N-myc downstream regulated 1 |
| chr6_+_18387570 | 1.28 |
ENST00000259939.3
|
RNF144B
|
ring finger protein 144B |
| chr2_-_214013353 | 1.23 |
ENST00000451136.2
ENST00000421754.2 ENST00000374327.4 ENST00000413091.3 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr4_-_102268708 | 1.22 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr5_+_166711804 | 1.19 |
ENST00000518659.1
ENST00000545108.1 |
TENM2
|
teneurin transmembrane protein 2 |
| chr12_-_106480587 | 1.11 |
ENST00000548902.1
|
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr3_-_168865522 | 1.08 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr6_-_56716686 | 1.06 |
ENST00000520645.1
|
DST
|
dystonin |
| chr12_-_29936731 | 1.04 |
ENST00000552618.1
ENST00000539277.1 ENST00000551659.1 |
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr21_+_39628852 | 1.03 |
ENST00000398938.2
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr1_+_220701456 | 0.98 |
ENST00000366918.4
ENST00000402574.1 |
MARK1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr2_+_28974531 | 0.98 |
ENST00000420282.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr1_-_109935819 | 0.95 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr10_-_25010795 | 0.94 |
ENST00000416305.1
ENST00000376410.2 |
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr1_-_228135599 | 0.85 |
ENST00000272164.5
|
WNT9A
|
wingless-type MMTV integration site family, member 9A |
| chr18_+_21033239 | 0.85 |
ENST00000581585.1
ENST00000577501.1 |
RIOK3
|
RIO kinase 3 |
| chr3_+_38032216 | 0.84 |
ENST00000416303.1
|
VILL
|
villin-like |
| chr4_-_74486347 | 0.81 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr5_-_127674883 | 0.80 |
ENST00000507835.1
|
FBN2
|
fibrillin 2 |
| chr13_+_52598827 | 0.79 |
ENST00000521776.2
|
UTP14C
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast) |
| chr6_+_15401075 | 0.77 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr12_-_31479107 | 0.77 |
ENST00000542983.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr4_-_102268484 | 0.76 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr10_+_112631547 | 0.74 |
ENST00000280154.7
ENST00000393104.2 |
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor) |
| chr11_-_46639436 | 0.74 |
ENST00000532281.1
|
HARBI1
|
harbinger transposase derived 1 |
| chr19_-_4831701 | 0.73 |
ENST00000248244.5
|
TICAM1
|
toll-like receptor adaptor molecule 1 |
| chr1_-_100231349 | 0.73 |
ENST00000287474.5
ENST00000414213.1 |
FRRS1
|
ferric-chelate reductase 1 |
| chr2_+_28974489 | 0.72 |
ENST00000455580.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr1_-_116383738 | 0.69 |
ENST00000320238.3
|
NHLH2
|
nescient helix loop helix 2 |
| chr10_+_114710425 | 0.68 |
ENST00000352065.5
ENST00000369395.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr21_+_39628655 | 0.67 |
ENST00000398925.1
ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr4_-_74486109 | 0.66 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr5_+_40841410 | 0.64 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr5_-_115872124 | 0.64 |
ENST00000515009.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr3_-_49131614 | 0.64 |
ENST00000450685.1
|
QRICH1
|
glutamine-rich 1 |
| chr2_-_208030647 | 0.64 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr11_+_10477733 | 0.61 |
ENST00000528723.1
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr3_-_71632894 | 0.61 |
ENST00000493089.1
|
FOXP1
|
forkhead box P1 |
| chr22_-_38699003 | 0.59 |
ENST00000451964.1
|
CSNK1E
|
casein kinase 1, epsilon |
| chr5_-_115872142 | 0.58 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr13_-_99667960 | 0.58 |
ENST00000448493.2
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr4_-_99578789 | 0.58 |
ENST00000511651.1
ENST00000505184.1 |
TSPAN5
|
tetraspanin 5 |
| chr20_+_9049303 | 0.56 |
ENST00000407043.2
ENST00000441846.1 |
PLCB4
|
phospholipase C, beta 4 |
| chr16_+_53920795 | 0.56 |
ENST00000431610.2
ENST00000460382.1 |
FTO
|
fat mass and obesity associated |
| chr1_-_149982624 | 0.56 |
ENST00000417191.1
ENST00000369135.4 |
OTUD7B
|
OTU domain containing 7B |
| chr17_-_48546232 | 0.55 |
ENST00000258969.4
|
CHAD
|
chondroadherin |
| chr6_+_45296048 | 0.54 |
ENST00000465038.2
ENST00000352853.5 ENST00000541979.1 ENST00000371438.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr5_+_135468516 | 0.53 |
ENST00000507118.1
ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5
|
SMAD family member 5 |
| chr1_+_39670423 | 0.53 |
ENST00000536367.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr22_+_41487711 | 0.53 |
ENST00000263253.7
|
EP300
|
E1A binding protein p300 |
| chr3_-_16524357 | 0.53 |
ENST00000432519.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr17_+_41363854 | 0.52 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr18_+_21032781 | 0.52 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr17_-_48546324 | 0.51 |
ENST00000508540.1
|
CHAD
|
chondroadherin |
| chr1_-_94079648 | 0.51 |
ENST00000370247.3
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr1_-_243349684 | 0.51 |
ENST00000522895.1
|
CEP170
|
centrosomal protein 170kDa |
| chr12_+_27849378 | 0.50 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chr1_-_28384598 | 0.50 |
ENST00000373864.1
|
EYA3
|
eyes absent homolog 3 (Drosophila) |
| chr15_+_74911430 | 0.49 |
ENST00000562670.1
ENST00000564096.1 |
CLK3
|
CDC-like kinase 3 |
| chr17_+_8924837 | 0.49 |
ENST00000173229.2
|
NTN1
|
netrin 1 |
| chr9_+_127054254 | 0.49 |
ENST00000422297.1
|
NEK6
|
NIMA-related kinase 6 |
| chr8_+_120079478 | 0.48 |
ENST00000332843.2
|
COLEC10
|
collectin sub-family member 10 (C-type lectin) |
| chrX_+_105936982 | 0.48 |
ENST00000418562.1
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr1_+_61542922 | 0.47 |
ENST00000407417.3
|
NFIA
|
nuclear factor I/A |
| chr6_+_47666275 | 0.46 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr13_+_98086445 | 0.44 |
ENST00000245304.4
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr5_-_131132658 | 0.44 |
ENST00000514667.1
ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3
FNIP1
|
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr2_-_207023918 | 0.43 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr5_+_40841276 | 0.43 |
ENST00000254691.5
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr12_-_116714564 | 0.43 |
ENST00000548743.1
|
MED13L
|
mediator complex subunit 13-like |
| chr11_-_8739566 | 0.43 |
ENST00000533020.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr3_-_187454281 | 0.43 |
ENST00000232014.4
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr2_-_207024134 | 0.43 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr12_-_12714025 | 0.42 |
ENST00000539940.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr22_+_46449674 | 0.42 |
ENST00000381051.2
|
FLJ27365
|
hsa-mir-4763 |
| chr6_+_45296391 | 0.42 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr21_+_39628780 | 0.42 |
ENST00000417042.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr5_-_36241900 | 0.42 |
ENST00000381937.4
ENST00000514504.1 |
NADK2
|
NAD kinase 2, mitochondrial |
| chr15_-_30114622 | 0.42 |
ENST00000495972.2
ENST00000346128.6 |
TJP1
|
tight junction protein 1 |
| chr1_+_12524965 | 0.42 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr4_+_71588372 | 0.42 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr6_+_108882069 | 0.42 |
ENST00000406360.1
|
FOXO3
|
forkhead box O3 |
| chr19_+_52848659 | 0.42 |
ENST00000327920.8
|
ZNF610
|
zinc finger protein 610 |
| chr11_+_10476851 | 0.41 |
ENST00000396553.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr12_+_99038919 | 0.41 |
ENST00000551964.1
|
APAF1
|
apoptotic peptidase activating factor 1 |
| chr16_+_4666475 | 0.41 |
ENST00000591895.1
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr2_+_210518057 | 0.41 |
ENST00000452717.1
|
MAP2
|
microtubule-associated protein 2 |
| chr4_-_99578776 | 0.40 |
ENST00000515287.1
|
TSPAN5
|
tetraspanin 5 |
| chr2_+_28974603 | 0.40 |
ENST00000441461.1
ENST00000358506.2 |
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr12_-_12714006 | 0.39 |
ENST00000541207.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr14_-_61191049 | 0.38 |
ENST00000556952.3
|
SIX4
|
SIX homeobox 4 |
| chr14_-_89960395 | 0.38 |
ENST00000555034.1
ENST00000553904.1 |
FOXN3
|
forkhead box N3 |
| chr13_-_36050819 | 0.37 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr4_-_153303658 | 0.37 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr17_+_17685422 | 0.37 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr4_-_147443043 | 0.37 |
ENST00000394059.4
ENST00000502607.1 ENST00000335472.7 ENST00000432059.2 ENST00000394062.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr3_+_57875711 | 0.37 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr5_-_124082279 | 0.37 |
ENST00000513986.1
|
ZNF608
|
zinc finger protein 608 |
| chr4_-_102268628 | 0.36 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr2_+_210517895 | 0.36 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr2_-_207024233 | 0.35 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr10_-_104913367 | 0.35 |
ENST00000423468.2
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr2_+_33359687 | 0.35 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr10_+_13652047 | 0.34 |
ENST00000601460.1
|
RP11-295P9.3
|
Uncharacterized protein |
| chr1_-_94312706 | 0.34 |
ENST00000370244.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr2_-_220042825 | 0.34 |
ENST00000409789.1
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr1_+_43855545 | 0.34 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr15_+_96869165 | 0.34 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr3_-_128902729 | 0.33 |
ENST00000451728.2
ENST00000446936.2 ENST00000502976.1 ENST00000500450.2 ENST00000441626.2 |
CNBP
|
CCHC-type zinc finger, nucleic acid binding protein |
| chr9_-_89562104 | 0.32 |
ENST00000298743.7
|
GAS1
|
growth arrest-specific 1 |
| chr4_+_57774042 | 0.32 |
ENST00000309042.7
|
REST
|
RE1-silencing transcription factor |
| chr9_-_136933615 | 0.31 |
ENST00000371834.2
|
BRD3
|
bromodomain containing 3 |
| chr22_-_43036607 | 0.31 |
ENST00000505920.1
|
ATP5L2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2 |
| chr2_-_8977714 | 0.31 |
ENST00000319688.5
ENST00000489024.1 ENST00000256707.3 ENST00000427284.1 ENST00000418530.1 ENST00000473731.1 |
KIDINS220
|
kinase D-interacting substrate, 220kDa |
| chr1_+_155099927 | 0.31 |
ENST00000368407.3
|
EFNA1
|
ephrin-A1 |
| chr2_+_172309634 | 0.31 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr5_-_36242119 | 0.31 |
ENST00000511088.1
ENST00000282512.3 ENST00000506945.1 |
NADK2
|
NAD kinase 2, mitochondrial |
| chr3_-_49131788 | 0.30 |
ENST00000395443.2
ENST00000411682.1 |
QRICH1
|
glutamine-rich 1 |
| chr10_+_114710516 | 0.30 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr16_+_67552303 | 0.30 |
ENST00000562116.1
|
FAM65A
|
family with sequence similarity 65, member A |
| chr8_+_11666649 | 0.29 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chrX_-_117107680 | 0.29 |
ENST00000447671.2
ENST00000262820.3 |
KLHL13
|
kelch-like family member 13 |
| chr2_+_181845532 | 0.29 |
ENST00000602475.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr5_+_179135246 | 0.29 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr3_-_133969673 | 0.28 |
ENST00000427044.2
|
RYK
|
receptor-like tyrosine kinase |
| chr2_+_33359646 | 0.28 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr11_-_8739383 | 0.28 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr6_-_52926539 | 0.27 |
ENST00000350082.5
ENST00000356971.3 |
ICK
|
intestinal cell (MAK-like) kinase |
| chr5_+_159614374 | 0.27 |
ENST00000393980.4
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr19_-_13900972 | 0.26 |
ENST00000397557.1
|
AC008686.1
|
Uncharacterized protein |
| chr1_+_174846570 | 0.26 |
ENST00000392064.2
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr2_+_28974668 | 0.26 |
ENST00000296122.6
ENST00000395366.2 |
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr12_+_93096759 | 0.26 |
ENST00000544406.2
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr11_+_27076764 | 0.26 |
ENST00000525090.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr12_-_122018114 | 0.25 |
ENST00000539394.1
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr9_+_127054217 | 0.25 |
ENST00000394199.2
ENST00000546191.1 |
NEK6
|
NIMA-related kinase 6 |
| chr1_+_39670360 | 0.24 |
ENST00000494012.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr5_-_39424961 | 0.24 |
ENST00000503513.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr1_+_43855560 | 0.24 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr3_-_195310802 | 0.24 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr19_+_3880581 | 0.24 |
ENST00000450849.2
ENST00000301260.6 ENST00000398448.3 |
ATCAY
|
ataxia, cerebellar, Cayman type |
| chr4_-_186696425 | 0.24 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_-_42418999 | 0.23 |
ENST00000340840.2
ENST00000354325.2 |
TRERF1
|
transcriptional regulating factor 1 |
| chr3_+_57875738 | 0.23 |
ENST00000417128.1
ENST00000438794.1 |
SLMAP
|
sarcolemma associated protein |
| chr8_-_116673894 | 0.23 |
ENST00000395713.2
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr8_-_114449112 | 0.23 |
ENST00000455883.2
ENST00000352409.3 ENST00000297405.5 |
CSMD3
|
CUB and Sushi multiple domains 3 |
| chr3_-_128902759 | 0.23 |
ENST00000422453.2
ENST00000504813.1 ENST00000512338.1 |
CNBP
|
CCHC-type zinc finger, nucleic acid binding protein |
| chr7_+_151038850 | 0.23 |
ENST00000355851.4
ENST00000566856.1 ENST00000470229.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr5_+_176692466 | 0.22 |
ENST00000508029.1
ENST00000503056.1 |
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr7_-_139876734 | 0.22 |
ENST00000006967.5
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr12_+_93115281 | 0.22 |
ENST00000549856.1
|
PLEKHG7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr8_-_71157595 | 0.22 |
ENST00000519724.1
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr12_-_53994805 | 0.22 |
ENST00000328463.7
|
ATF7
|
activating transcription factor 7 |
| chr3_-_141868357 | 0.22 |
ENST00000489671.1
ENST00000475734.1 ENST00000467072.1 ENST00000499676.2 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr4_+_71587669 | 0.21 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr12_+_10365082 | 0.21 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr20_+_44509857 | 0.21 |
ENST00000372523.1
ENST00000372520.1 |
ZSWIM1
|
zinc finger, SWIM-type containing 1 |
| chr12_+_10365404 | 0.21 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr2_+_33359473 | 0.21 |
ENST00000432635.1
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr7_-_139477500 | 0.21 |
ENST00000406875.3
ENST00000428878.2 |
HIPK2
|
homeodomain interacting protein kinase 2 |
| chr4_+_28437071 | 0.20 |
ENST00000509416.1
|
RP11-123O22.1
|
RP11-123O22.1 |
| chrX_+_22050546 | 0.20 |
ENST00000379374.4
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr18_+_32073253 | 0.19 |
ENST00000283365.9
ENST00000596745.1 ENST00000315456.6 |
DTNA
|
dystrobrevin, alpha |
| chr4_-_83765613 | 0.19 |
ENST00000503937.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr3_-_141868293 | 0.18 |
ENST00000317104.7
ENST00000494358.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr12_+_93096619 | 0.18 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr8_-_141931287 | 0.18 |
ENST00000517887.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr2_+_181845763 | 0.18 |
ENST00000602499.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr3_-_24207039 | 0.17 |
ENST00000280696.5
|
THRB
|
thyroid hormone receptor, beta |
| chr2_-_207078154 | 0.17 |
ENST00000447845.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr12_-_49453557 | 0.17 |
ENST00000547610.1
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr1_+_22778337 | 0.17 |
ENST00000404138.1
ENST00000400239.2 ENST00000375647.4 ENST00000374651.4 |
ZBTB40
|
zinc finger and BTB domain containing 40 |
| chr6_-_134639042 | 0.17 |
ENST00000461976.2
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr5_-_39425222 | 0.17 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr20_+_58203664 | 0.17 |
ENST00000541461.1
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr7_+_151038785 | 0.16 |
ENST00000413040.2
ENST00000568733.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr3_-_15382875 | 0.16 |
ENST00000408919.3
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr3_-_134092561 | 0.16 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr9_+_71944241 | 0.16 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr10_+_114710211 | 0.16 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr8_-_23261589 | 0.16 |
ENST00000524168.1
ENST00000523833.2 ENST00000519243.1 ENST00000389131.3 |
LOXL2
|
lysyl oxidase-like 2 |
| chr18_-_53070913 | 0.16 |
ENST00000568186.1
ENST00000564228.1 |
TCF4
|
transcription factor 4 |
| chr5_+_65222500 | 0.16 |
ENST00000511297.1
ENST00000506030.1 |
ERBB2IP
|
erbb2 interacting protein |
| chrX_-_117107542 | 0.16 |
ENST00000371878.1
|
KLHL13
|
kelch-like family member 13 |
| chr11_-_108464465 | 0.16 |
ENST00000525344.1
|
EXPH5
|
exophilin 5 |
| chr17_+_71229346 | 0.15 |
ENST00000535032.2
ENST00000582793.1 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr18_+_19749386 | 0.15 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr1_-_153931052 | 0.15 |
ENST00000368630.3
ENST00000368633.1 |
CRTC2
|
CREB regulated transcription coactivator 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXC1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.7 | 2.2 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.5 | 5.5 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.5 | 2.0 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.4 | 2.5 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.3 | 1.4 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.3 | 1.1 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.2 | 2.7 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.2 | 0.8 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.2 | 1.7 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 0.6 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 1.6 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 1.0 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 1.2 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.3 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.1 | 0.4 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.9 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.5 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 | 2.4 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 1.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.4 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 | 0.4 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 0.3 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.4 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 0.6 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.6 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.9 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 1.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 0.7 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.3 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.6 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.3 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 0.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.4 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.2 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 0.7 | GO:0045359 | positive regulation of chemokine biosynthetic process(GO:0045080) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.1 | 0.6 | GO:1902510 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.1 | 1.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.5 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.2 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.2 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 | 0.3 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 0.3 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.1 | 1.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.8 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 0.2 | GO:0007493 | endodermal cell fate determination(GO:0007493) |
| 0.0 | 1.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.3 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.3 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) notochord formation(GO:0014028) |
| 0.0 | 0.5 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 0.0 | 0.2 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.4 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.8 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.2 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.2 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.5 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 2.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.9 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.2 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.2 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 1.1 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.7 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.9 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.2 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 1.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.3 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0002249 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.8 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 1.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.9 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.6 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.2 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.0 | 0.8 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.2 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 2.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.5 | 4.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 1.2 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.2 | 1.1 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 2.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 1.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.6 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 1.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.6 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.7 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.7 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 1.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.7 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.9 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 2.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 2.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.6 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.5 | 2.4 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.3 | 4.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 0.9 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.6 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.1 | 0.8 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 2.0 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.7 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.4 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.1 | 1.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.4 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 2.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.8 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.3 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 0.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 2.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 1.2 | GO:0070016 | gamma-catenin binding(GO:0045295) armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 5.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.7 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 1.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.6 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.4 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 1.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 1.0 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.7 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 1.3 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.4 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.4 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 4.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 2.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 3.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 2.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 2.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.7 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 2.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.7 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 1.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 5.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 1.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |