Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
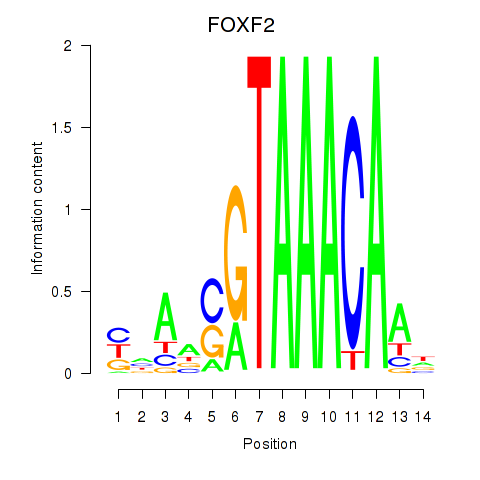
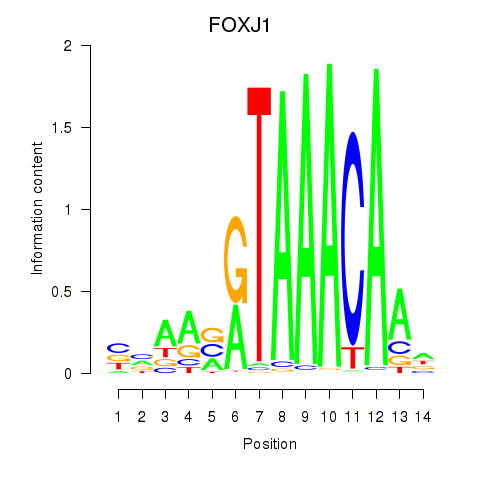
Results for FOXF2_FOXJ1
Z-value: 0.23
Transcription factors associated with FOXF2_FOXJ1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXF2
|
ENSG00000137273.3 | forkhead box F2 |
|
FOXJ1
|
ENSG00000129654.7 | forkhead box J1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXJ1 | hg19_v2_chr17_-_74137374_74137385 | -0.22 | 3.6e-01 | Click! |
| FOXF2 | hg19_v2_chr6_+_1389989_1390069 | 0.22 | 3.6e-01 | Click! |
Activity profile of FOXF2_FOXJ1 motif
Sorted Z-values of FOXF2_FOXJ1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_77233294 | 0.32 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr19_+_18496957 | 0.29 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr4_-_140223614 | 0.27 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr17_+_72427477 | 0.26 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr4_-_140223670 | 0.25 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr2_-_233877912 | 0.23 |
ENST00000264051.3
|
NGEF
|
neuronal guanine nucleotide exchange factor |
| chr3_-_48956818 | 0.22 |
ENST00000408959.2
|
ARIH2OS
|
ariadne homolog 2 opposite strand |
| chr6_+_21666633 | 0.22 |
ENST00000606851.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr12_-_53343602 | 0.22 |
ENST00000546897.1
ENST00000552551.1 |
KRT8
|
keratin 8 |
| chr17_+_67410832 | 0.21 |
ENST00000590474.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr17_+_72426891 | 0.20 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr11_+_844406 | 0.20 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr11_-_115375107 | 0.19 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr2_-_165424973 | 0.18 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr11_+_844067 | 0.18 |
ENST00000397406.1
ENST00000409543.2 ENST00000525201.1 |
TSPAN4
|
tetraspanin 4 |
| chr8_+_52730143 | 0.18 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr2_+_33661382 | 0.17 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr10_+_70847852 | 0.16 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr19_+_50380917 | 0.16 |
ENST00000535102.2
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr2_-_157198860 | 0.15 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr19_-_36523709 | 0.15 |
ENST00000592017.1
ENST00000360535.4 |
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr11_-_67374177 | 0.15 |
ENST00000333139.3
|
C11orf72
|
chromosome 11 open reading frame 72 |
| chr17_+_16284604 | 0.14 |
ENST00000395839.1
ENST00000395837.1 |
UBB
|
ubiquitin B |
| chr19_+_50380682 | 0.14 |
ENST00000221543.5
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr20_-_22566089 | 0.14 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr12_-_92539614 | 0.14 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr9_+_136287444 | 0.13 |
ENST00000355699.2
ENST00000356589.2 ENST00000371911.3 |
ADAMTS13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr3_-_58523010 | 0.13 |
ENST00000459701.2
ENST00000302819.5 |
ACOX2
|
acyl-CoA oxidase 2, branched chain |
| chr12_-_53343633 | 0.13 |
ENST00000546826.1
|
KRT8
|
keratin 8 |
| chr18_-_56296182 | 0.13 |
ENST00000361673.3
|
ALPK2
|
alpha-kinase 2 |
| chr20_-_62582475 | 0.13 |
ENST00000369908.5
|
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr15_+_63188009 | 0.13 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr1_+_66796401 | 0.12 |
ENST00000528771.1
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr12_-_53343560 | 0.12 |
ENST00000548998.1
|
KRT8
|
keratin 8 |
| chr16_+_777739 | 0.12 |
ENST00000563792.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr3_+_50712672 | 0.12 |
ENST00000266037.9
|
DOCK3
|
dedicator of cytokinesis 3 |
| chr19_-_36523529 | 0.11 |
ENST00000593074.1
|
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr12_+_20963632 | 0.11 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr12_+_20963647 | 0.11 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr9_-_4859260 | 0.10 |
ENST00000599351.1
|
AL158147.2
|
HCG2011465; Uncharacterized protein |
| chr14_+_23299088 | 0.10 |
ENST00000355151.5
ENST00000397496.3 ENST00000555345.1 ENST00000432849.3 ENST00000553711.1 ENST00000556465.1 ENST00000397505.2 ENST00000557221.1 ENST00000311892.6 ENST00000556840.1 ENST00000555536.1 |
MRPL52
|
mitochondrial ribosomal protein L52 |
| chr1_-_246729544 | 0.10 |
ENST00000544618.1
ENST00000366514.4 |
TFB2M
|
transcription factor B2, mitochondrial |
| chr6_+_84563295 | 0.10 |
ENST00000369687.1
|
RIPPLY2
|
ripply transcriptional repressor 2 |
| chr16_+_28763108 | 0.10 |
ENST00000357796.3
ENST00000550983.1 |
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr17_-_26662464 | 0.09 |
ENST00000579419.1
ENST00000585313.1 ENST00000395418.3 ENST00000578985.1 ENST00000577498.1 ENST00000585089.1 ENST00000357896.3 |
IFT20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr22_-_37505449 | 0.09 |
ENST00000406725.1
|
TMPRSS6
|
transmembrane protease, serine 6 |
| chr1_+_185703513 | 0.09 |
ENST00000271588.4
ENST00000367492.2 |
HMCN1
|
hemicentin 1 |
| chr1_+_22351977 | 0.09 |
ENST00000420503.1
ENST00000416769.1 ENST00000404210.2 |
LINC00339
|
long intergenic non-protein coding RNA 339 |
| chr19_+_18492973 | 0.09 |
ENST00000595973.2
|
GDF15
|
growth differentiation factor 15 |
| chr15_+_74509530 | 0.09 |
ENST00000321288.5
|
CCDC33
|
coiled-coil domain containing 33 |
| chr16_+_777246 | 0.09 |
ENST00000561546.1
ENST00000564545.1 ENST00000389703.3 ENST00000567414.1 ENST00000568141.1 |
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr10_-_375422 | 0.09 |
ENST00000434695.2
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr16_-_28374829 | 0.09 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family, member B6 |
| chr7_-_37026108 | 0.09 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr8_+_35649365 | 0.09 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr14_-_54418598 | 0.09 |
ENST00000609748.1
ENST00000558961.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr2_+_66918558 | 0.09 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr20_+_44035200 | 0.09 |
ENST00000372717.1
ENST00000360981.4 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr16_-_86542455 | 0.08 |
ENST00000595886.1
ENST00000597578.1 ENST00000593604.1 |
FENDRR
|
FOXF1 adjacent non-coding developmental regulatory RNA |
| chr3_-_185826855 | 0.08 |
ENST00000306376.5
|
ETV5
|
ets variant 5 |
| chr9_+_78505581 | 0.08 |
ENST00000376767.3
ENST00000376752.4 |
PCSK5
|
proprotein convertase subtilisin/kexin type 5 |
| chr17_-_6947225 | 0.08 |
ENST00000574600.1
ENST00000308009.1 ENST00000447225.1 |
SLC16A11
|
solute carrier family 16, member 11 |
| chr22_-_37505588 | 0.08 |
ENST00000406856.1
|
TMPRSS6
|
transmembrane protease, serine 6 |
| chr2_+_58655461 | 0.08 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr4_+_124571409 | 0.08 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr7_-_25702669 | 0.08 |
ENST00000446840.1
|
AC003090.1
|
AC003090.1 |
| chr6_-_25830785 | 0.08 |
ENST00000468082.1
|
SLC17A1
|
solute carrier family 17 (organic anion transporter), member 1 |
| chr7_+_134832808 | 0.08 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr2_+_233527443 | 0.07 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr12_-_96390108 | 0.07 |
ENST00000538703.1
ENST00000261208.3 |
HAL
|
histidine ammonia-lyase |
| chr10_-_13570533 | 0.07 |
ENST00000396900.2
ENST00000396898.2 |
BEND7
|
BEN domain containing 7 |
| chr10_-_73848086 | 0.07 |
ENST00000536168.1
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr4_-_123542224 | 0.07 |
ENST00000264497.3
|
IL21
|
interleukin 21 |
| chr15_-_76352069 | 0.07 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr12_-_56352368 | 0.07 |
ENST00000549404.1
|
PMEL
|
premelanosome protein |
| chr2_+_61404545 | 0.07 |
ENST00000357022.2
|
AHSA2
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) |
| chr17_-_8263538 | 0.06 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr11_-_63933504 | 0.06 |
ENST00000255681.6
|
MACROD1
|
MACRO domain containing 1 |
| chr8_+_97597148 | 0.06 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr2_+_74120094 | 0.06 |
ENST00000409731.3
ENST00000345517.3 ENST00000409918.1 ENST00000442912.1 ENST00000409624.1 |
ACTG2
|
actin, gamma 2, smooth muscle, enteric |
| chr14_+_105267250 | 0.06 |
ENST00000342537.7
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr4_-_174256276 | 0.06 |
ENST00000296503.5
|
HMGB2
|
high mobility group box 2 |
| chr13_-_99910673 | 0.06 |
ENST00000397473.2
ENST00000397470.2 |
GPR18
|
G protein-coupled receptor 18 |
| chr10_+_52751010 | 0.06 |
ENST00000373985.1
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr11_-_27723158 | 0.06 |
ENST00000395980.2
|
BDNF
|
brain-derived neurotrophic factor |
| chr2_+_58655520 | 0.06 |
ENST00000455219.3
ENST00000449448.2 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr5_-_180076613 | 0.06 |
ENST00000261937.6
ENST00000393347.3 |
FLT4
|
fms-related tyrosine kinase 4 |
| chr12_-_96389702 | 0.06 |
ENST00000552509.1
|
HAL
|
histidine ammonia-lyase |
| chr5_+_140529630 | 0.06 |
ENST00000543635.1
|
PCDHB6
|
protocadherin beta 6 |
| chr2_-_39456673 | 0.06 |
ENST00000378803.1
ENST00000395035.3 |
CDKL4
|
cyclin-dependent kinase-like 4 |
| chr3_+_98451532 | 0.06 |
ENST00000486334.2
ENST00000394162.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr2_+_232575168 | 0.06 |
ENST00000440384.1
|
PTMA
|
prothymosin, alpha |
| chr1_+_150122034 | 0.06 |
ENST00000025469.6
ENST00000369124.4 |
PLEKHO1
|
pleckstrin homology domain containing, family O member 1 |
| chr6_-_32122106 | 0.06 |
ENST00000428778.1
|
PRRT1
|
proline-rich transmembrane protein 1 |
| chr17_+_22022437 | 0.06 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr8_-_145754428 | 0.06 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr20_-_52645231 | 0.06 |
ENST00000448484.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr13_-_99910620 | 0.06 |
ENST00000416594.1
|
GPR18
|
G protein-coupled receptor 18 |
| chr12_+_28605426 | 0.06 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr1_+_25664408 | 0.06 |
ENST00000374358.4
|
TMEM50A
|
transmembrane protein 50A |
| chr20_+_44420570 | 0.06 |
ENST00000372622.3
|
DNTTIP1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr19_-_49016847 | 0.06 |
ENST00000598924.1
|
CTC-273B12.10
|
CTC-273B12.10 |
| chr8_-_133772794 | 0.06 |
ENST00000519187.1
ENST00000523829.1 ENST00000356838.3 ENST00000377901.4 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chr1_+_149239529 | 0.06 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr17_-_7082668 | 0.06 |
ENST00000573083.1
ENST00000574388.1 |
ASGR1
|
asialoglycoprotein receptor 1 |
| chr4_+_117220016 | 0.06 |
ENST00000604093.1
|
MTRNR2L13
|
MT-RNR2-like 13 (pseudogene) |
| chr12_+_50135588 | 0.06 |
ENST00000423828.1
ENST00000550445.1 |
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr6_+_12717892 | 0.06 |
ENST00000379350.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr2_+_162272605 | 0.06 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr20_-_44993012 | 0.06 |
ENST00000372229.1
ENST00000372230.5 ENST00000543605.1 ENST00000243896.2 ENST00000317734.8 |
SLC35C2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr2_+_61404624 | 0.05 |
ENST00000394457.3
|
AHSA2
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) |
| chr6_+_22221010 | 0.05 |
ENST00000567753.1
|
RP11-524C21.2
|
RP11-524C21.2 |
| chr4_+_15376165 | 0.05 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr19_-_50380536 | 0.05 |
ENST00000391832.3
ENST00000391834.2 ENST00000344175.5 |
AKT1S1
|
AKT1 substrate 1 (proline-rich) |
| chr8_+_31497271 | 0.05 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr15_+_28624878 | 0.05 |
ENST00000450328.2
|
GOLGA8F
|
golgin A8 family, member F |
| chr8_+_67344710 | 0.05 |
ENST00000379385.4
ENST00000396623.3 ENST00000415254.1 |
ADHFE1
|
alcohol dehydrogenase, iron containing, 1 |
| chr18_+_77160282 | 0.05 |
ENST00000318065.5
ENST00000545796.1 ENST00000592223.1 ENST00000329101.4 ENST00000586434.1 |
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr1_+_15668240 | 0.05 |
ENST00000444385.1
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr11_+_71938925 | 0.05 |
ENST00000538751.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr9_-_130679257 | 0.05 |
ENST00000361444.3
ENST00000335791.5 ENST00000343609.2 |
ST6GALNAC4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr19_-_49016418 | 0.05 |
ENST00000270238.3
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr8_-_133772870 | 0.05 |
ENST00000522334.1
ENST00000519016.1 |
TMEM71
|
transmembrane protein 71 |
| chr8_-_116673894 | 0.05 |
ENST00000395713.2
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr2_-_102091144 | 0.05 |
ENST00000428343.1
|
RFX8
|
RFX family member 8, lacking RFX DNA binding domain |
| chr3_+_171561127 | 0.05 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr4_+_166794383 | 0.05 |
ENST00000061240.2
ENST00000507499.1 |
TLL1
|
tolloid-like 1 |
| chr5_-_180076580 | 0.05 |
ENST00000502649.1
|
FLT4
|
fms-related tyrosine kinase 4 |
| chr11_-_85397167 | 0.05 |
ENST00000316398.3
|
CCDC89
|
coiled-coil domain containing 89 |
| chr18_+_3449330 | 0.05 |
ENST00000549253.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr19_+_13906250 | 0.05 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr11_-_118972575 | 0.05 |
ENST00000432443.2
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr22_-_39928823 | 0.05 |
ENST00000334678.3
|
RPS19BP1
|
ribosomal protein S19 binding protein 1 |
| chr5_+_176853702 | 0.05 |
ENST00000507633.1
ENST00000393576.3 ENST00000355958.5 ENST00000528793.1 ENST00000512684.1 |
GRK6
|
G protein-coupled receptor kinase 6 |
| chr17_-_26662440 | 0.04 |
ENST00000578122.1
|
IFT20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr5_+_176853669 | 0.04 |
ENST00000355472.5
|
GRK6
|
G protein-coupled receptor kinase 6 |
| chr6_+_101846664 | 0.04 |
ENST00000421544.1
ENST00000413795.1 ENST00000369138.1 ENST00000358361.3 |
GRIK2
|
glutamate receptor, ionotropic, kainate 2 |
| chr9_+_113431059 | 0.04 |
ENST00000416899.2
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase |
| chr14_+_32964258 | 0.04 |
ENST00000556638.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr5_-_36301984 | 0.04 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chrX_-_100129320 | 0.04 |
ENST00000372966.3
|
NOX1
|
NADPH oxidase 1 |
| chr11_+_113930955 | 0.04 |
ENST00000535700.1
|
ZBTB16
|
zinc finger and BTB domain containing 16 |
| chr2_+_105050794 | 0.04 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr5_-_150460914 | 0.04 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr12_-_63328817 | 0.04 |
ENST00000228705.6
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr4_+_24661479 | 0.04 |
ENST00000569621.1
|
RP11-496D24.2
|
RP11-496D24.2 |
| chr9_-_86432547 | 0.04 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr1_-_24469602 | 0.04 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1 |
| chr18_+_18943554 | 0.04 |
ENST00000580732.2
|
GREB1L
|
growth regulation by estrogen in breast cancer-like |
| chr8_+_99956662 | 0.04 |
ENST00000523368.1
ENST00000297565.4 ENST00000435298.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr6_-_43027105 | 0.04 |
ENST00000230413.5
ENST00000487429.1 ENST00000489623.1 ENST00000468957.1 |
MRPL2
|
mitochondrial ribosomal protein L2 |
| chr20_+_44420617 | 0.04 |
ENST00000449078.1
ENST00000456939.1 |
DNTTIP1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr12_-_96390063 | 0.04 |
ENST00000541929.1
|
HAL
|
histidine ammonia-lyase |
| chr1_-_246357029 | 0.04 |
ENST00000391836.2
|
SMYD3
|
SET and MYND domain containing 3 |
| chr7_+_119913688 | 0.04 |
ENST00000331113.4
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr3_+_132316081 | 0.04 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr8_-_6420930 | 0.04 |
ENST00000325203.5
|
ANGPT2
|
angiopoietin 2 |
| chr2_-_220264703 | 0.04 |
ENST00000519905.1
ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr17_-_37934466 | 0.04 |
ENST00000583368.1
|
IKZF3
|
IKAROS family zinc finger 3 (Aiolos) |
| chr6_+_32121908 | 0.04 |
ENST00000375143.2
ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr12_-_58240470 | 0.04 |
ENST00000548823.1
ENST00000398073.2 |
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr11_-_5526834 | 0.03 |
ENST00000380237.1
ENST00000396895.1 ENST00000380252.1 |
HBE1
HBG2
|
hemoglobin, epsilon 1 hemoglobin, gamma G |
| chr22_-_39268308 | 0.03 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr6_-_42016385 | 0.03 |
ENST00000502771.1
ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3
|
cyclin D3 |
| chr19_+_2270283 | 0.03 |
ENST00000588673.2
|
OAZ1
|
ornithine decarboxylase antizyme 1 |
| chr8_-_6420759 | 0.03 |
ENST00000523120.1
|
ANGPT2
|
angiopoietin 2 |
| chr11_-_34535297 | 0.03 |
ENST00000532417.1
|
ELF5
|
E74-like factor 5 (ets domain transcription factor) |
| chr3_+_137717571 | 0.03 |
ENST00000343735.4
|
CLDN18
|
claudin 18 |
| chr12_-_772901 | 0.03 |
ENST00000305108.4
|
NINJ2
|
ninjurin 2 |
| chr17_+_26662730 | 0.03 |
ENST00000226225.2
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr17_+_58755184 | 0.03 |
ENST00000589222.1
ENST00000407086.3 ENST00000390652.5 |
BCAS3
|
breast carcinoma amplified sequence 3 |
| chr6_+_112375462 | 0.03 |
ENST00000361714.1
|
WISP3
|
WNT1 inducible signaling pathway protein 3 |
| chr8_-_6420777 | 0.03 |
ENST00000415216.1
|
ANGPT2
|
angiopoietin 2 |
| chr2_+_103236004 | 0.03 |
ENST00000233969.2
|
SLC9A2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr14_-_101036119 | 0.03 |
ENST00000355173.2
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr16_+_11439286 | 0.03 |
ENST00000312499.5
ENST00000576027.1 |
RMI2
|
RecQ mediated genome instability 2 |
| chr4_+_71108300 | 0.03 |
ENST00000304954.3
|
CSN3
|
casein kappa |
| chr15_+_72978521 | 0.03 |
ENST00000542334.1
ENST00000268057.4 |
BBS4
|
Bardet-Biedl syndrome 4 |
| chr6_+_32121789 | 0.03 |
ENST00000437001.2
ENST00000375137.2 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr20_+_33563206 | 0.03 |
ENST00000262873.7
|
MYH7B
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr8_+_118147498 | 0.03 |
ENST00000519688.1
ENST00000456015.2 |
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chr12_-_49504449 | 0.03 |
ENST00000547675.1
|
LMBR1L
|
limb development membrane protein 1-like |
| chr11_+_71791359 | 0.03 |
ENST00000419228.1
ENST00000435085.1 ENST00000307198.7 ENST00000538413.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr7_+_116593292 | 0.03 |
ENST00000393446.2
ENST00000265437.5 ENST00000393451.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr18_+_28898052 | 0.03 |
ENST00000257192.4
|
DSG1
|
desmoglein 1 |
| chr1_+_74701062 | 0.03 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr6_-_76203454 | 0.03 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr7_+_129906660 | 0.03 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr10_-_4285923 | 0.03 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr7_-_140340576 | 0.03 |
ENST00000275884.6
ENST00000475837.1 |
DENND2A
|
DENN/MADD domain containing 2A |
| chr17_-_40134339 | 0.03 |
ENST00000587727.1
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr17_-_29641104 | 0.03 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr15_-_40600111 | 0.03 |
ENST00000543785.2
ENST00000260402.3 |
PLCB2
|
phospholipase C, beta 2 |
| chr14_-_80782219 | 0.03 |
ENST00000553594.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr15_-_72978490 | 0.02 |
ENST00000311755.3
|
HIGD2B
|
HIG1 hypoxia inducible domain family, member 2B |
| chr17_-_29641084 | 0.02 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr4_-_186456652 | 0.02 |
ENST00000284767.5
ENST00000284770.5 |
PDLIM3
|
PDZ and LIM domain 3 |
| chr4_-_70518941 | 0.02 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr6_-_76203345 | 0.02 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr10_-_4285835 | 0.02 |
ENST00000454470.1
|
LINC00702
|
long intergenic non-protein coding RNA 702 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXF2_FOXJ1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.2 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.2 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.5 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.1 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.0 | 0.1 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.1 | GO:0060503 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.1 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.0 | 0.3 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0036328 | VEGF-C-activated receptor activity(GO:0036328) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.0 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |