Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
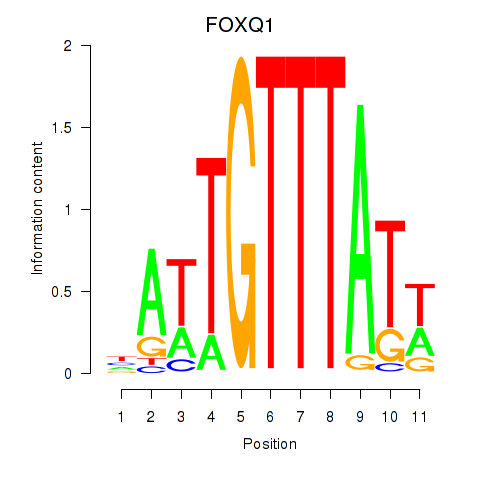
Results for FOXQ1
Z-value: 0.95
Transcription factors associated with FOXQ1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXQ1
|
ENSG00000164379.4 | forkhead box Q1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXQ1 | hg19_v2_chr6_+_1312675_1312701 | 0.10 | 6.8e-01 | Click! |
Activity profile of FOXQ1 motif
Sorted Z-values of FOXQ1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_193853927 | 2.96 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr18_+_61254570 | 2.76 |
ENST00000344731.5
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr7_-_41742697 | 2.68 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr10_-_98031265 | 2.60 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr1_-_182360918 | 2.46 |
ENST00000339526.4
|
GLUL
|
glutamate-ammonia ligase |
| chr18_+_61254534 | 2.40 |
ENST00000269489.5
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr1_-_182360498 | 2.31 |
ENST00000417584.2
|
GLUL
|
glutamate-ammonia ligase |
| chr18_+_61445007 | 2.19 |
ENST00000447428.1
ENST00000546027.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr18_+_61143994 | 2.19 |
ENST00000382771.4
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr8_-_124553437 | 2.19 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr4_-_119274121 | 2.12 |
ENST00000296498.3
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr18_+_61254221 | 2.08 |
ENST00000431153.1
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr10_-_98031310 | 2.03 |
ENST00000427367.2
ENST00000413476.2 |
BLNK
|
B-cell linker |
| chr8_-_49834299 | 1.95 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr8_-_49833978 | 1.94 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chrX_+_105936982 | 1.87 |
ENST00000418562.1
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr1_+_153003671 | 1.81 |
ENST00000307098.4
|
SPRR1B
|
small proline-rich protein 1B |
| chr10_-_10836919 | 1.73 |
ENST00000602763.1
ENST00000415590.2 ENST00000434919.2 |
SFTA1P
|
surfactant associated 1, pseudogene |
| chr1_+_70820451 | 1.48 |
ENST00000361764.4
ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3
|
HERV-H LTR-associating 3 |
| chr1_+_24645865 | 1.46 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_+_24645807 | 1.45 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr6_+_106988986 | 1.44 |
ENST00000457437.1
ENST00000535438.1 |
AIM1
|
absent in melanoma 1 |
| chr11_-_102668879 | 1.43 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr3_+_177159695 | 1.42 |
ENST00000442937.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr4_-_68749745 | 1.42 |
ENST00000283916.6
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr11_+_34654011 | 1.36 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr2_+_47596287 | 1.35 |
ENST00000263735.4
|
EPCAM
|
epithelial cell adhesion molecule |
| chr14_+_24867992 | 1.35 |
ENST00000382554.3
|
NYNRIN
|
NYN domain and retroviral integrase containing |
| chr1_+_24646002 | 1.31 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr3_+_136649311 | 1.27 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr3_+_189349162 | 1.24 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr1_+_26605618 | 1.22 |
ENST00000270792.5
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr20_+_12989596 | 1.21 |
ENST00000434210.1
ENST00000399002.2 |
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr5_-_24645078 | 1.21 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr14_-_81902791 | 1.20 |
ENST00000557055.1
|
STON2
|
stonin 2 |
| chr1_+_24645915 | 1.19 |
ENST00000350501.5
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chrM_+_10053 | 1.18 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr3_-_107777208 | 1.12 |
ENST00000398258.3
|
CD47
|
CD47 molecule |
| chr6_+_41604747 | 1.12 |
ENST00000419164.1
ENST00000373051.2 |
MDFI
|
MyoD family inhibitor |
| chr4_-_68749699 | 1.09 |
ENST00000545541.1
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr19_+_7580103 | 1.08 |
ENST00000596712.1
|
ZNF358
|
zinc finger protein 358 |
| chr21_+_39628852 | 1.06 |
ENST00000398938.2
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr3_+_182983090 | 1.05 |
ENST00000465010.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr1_+_174844645 | 1.04 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr6_+_30856507 | 1.00 |
ENST00000513240.1
ENST00000424544.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr3_-_12587055 | 0.99 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr4_+_69313145 | 0.98 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr13_+_76413852 | 0.98 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr1_+_24646263 | 0.98 |
ENST00000524724.1
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr4_-_159094194 | 0.97 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr6_-_28973037 | 0.97 |
ENST00000377179.3
|
ZNF311
|
zinc finger protein 311 |
| chr14_+_37126765 | 0.97 |
ENST00000402703.2
|
PAX9
|
paired box 9 |
| chr16_+_2820912 | 0.96 |
ENST00000570539.1
|
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr4_-_90756769 | 0.94 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr17_-_73851285 | 0.92 |
ENST00000589642.1
ENST00000593002.1 ENST00000590221.1 ENST00000344296.4 ENST00000587374.1 ENST00000585462.1 ENST00000433525.2 ENST00000254806.3 |
WBP2
|
WW domain binding protein 2 |
| chr4_-_80994210 | 0.92 |
ENST00000403729.2
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr1_+_104159999 | 0.92 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr4_-_80994619 | 0.91 |
ENST00000404191.1
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr2_+_158114051 | 0.89 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr5_+_65440032 | 0.88 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr16_-_4852915 | 0.86 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr11_-_117800080 | 0.86 |
ENST00000524993.1
ENST00000528626.1 ENST00000445164.2 ENST00000430170.2 ENST00000526090.1 |
TMPRSS13
|
transmembrane protease, serine 13 |
| chr15_+_49715449 | 0.85 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr8_+_79428539 | 0.81 |
ENST00000352966.5
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr10_+_75668916 | 0.81 |
ENST00000481390.1
|
PLAU
|
plasminogen activator, urokinase |
| chr10_+_5454505 | 0.80 |
ENST00000355029.4
|
NET1
|
neuroepithelial cell transforming 1 |
| chr5_+_102201722 | 0.80 |
ENST00000274392.9
ENST00000455264.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr6_+_131958436 | 0.79 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr6_-_113953705 | 0.78 |
ENST00000452675.1
|
RP11-367G18.1
|
RP11-367G18.1 |
| chr10_-_101380121 | 0.78 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr18_+_11752783 | 0.77 |
ENST00000585642.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr3_-_196911002 | 0.76 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr11_+_45943169 | 0.76 |
ENST00000529052.1
ENST00000531526.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chrM_+_10758 | 0.75 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr6_+_116575329 | 0.74 |
ENST00000430252.2
ENST00000540275.1 ENST00000448740.2 |
DSE
RP3-486I3.7
|
dermatan sulfate epimerase RP3-486I3.7 |
| chr3_+_122513642 | 0.73 |
ENST00000261038.5
|
DIRC2
|
disrupted in renal carcinoma 2 |
| chr19_-_46148820 | 0.71 |
ENST00000587152.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chrX_+_9880412 | 0.71 |
ENST00000418909.2
|
SHROOM2
|
shroom family member 2 |
| chr8_+_120079478 | 0.71 |
ENST00000332843.2
|
COLEC10
|
collectin sub-family member 10 (C-type lectin) |
| chr1_-_202927490 | 0.70 |
ENST00000340990.5
|
ADIPOR1
|
adiponectin receptor 1 |
| chr13_-_26795840 | 0.69 |
ENST00000381570.3
ENST00000399762.2 ENST00000346166.3 |
RNF6
|
ring finger protein (C3H2C3 type) 6 |
| chr10_+_5488564 | 0.68 |
ENST00000449083.1
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chr10_+_127661942 | 0.67 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr3_+_177159744 | 0.67 |
ENST00000439009.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr14_+_52456193 | 0.67 |
ENST00000261700.3
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr4_-_80994471 | 0.66 |
ENST00000295465.4
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr14_+_52456327 | 0.66 |
ENST00000556760.1
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr5_+_140743859 | 0.64 |
ENST00000518069.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr4_-_23735183 | 0.62 |
ENST00000507666.1
|
RP11-380P13.2
|
RP11-380P13.2 |
| chr7_+_44646218 | 0.62 |
ENST00000444676.1
ENST00000222673.5 |
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr6_-_138833630 | 0.62 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr12_-_54582655 | 0.61 |
ENST00000504338.1
ENST00000514685.1 ENST00000504797.1 ENST00000513838.1 ENST00000505128.1 ENST00000337581.3 ENST00000503306.1 ENST00000243112.5 ENST00000514196.1 ENST00000506169.1 ENST00000507904.1 ENST00000508394.2 |
SMUG1
|
single-strand-selective monofunctional uracil-DNA glycosylase 1 |
| chrX_-_50557302 | 0.60 |
ENST00000289292.7
|
SHROOM4
|
shroom family member 4 |
| chr4_+_75174180 | 0.60 |
ENST00000413830.1
|
EPGN
|
epithelial mitogen |
| chr20_+_56964169 | 0.60 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr17_+_45771420 | 0.60 |
ENST00000578982.1
|
TBKBP1
|
TBK1 binding protein 1 |
| chr15_+_84906338 | 0.59 |
ENST00000512109.1
|
GOLGA6L4
|
golgin A6 family-like 4 |
| chrX_+_105855160 | 0.59 |
ENST00000372544.2
ENST00000372548.4 |
CXorf57
|
chromosome X open reading frame 57 |
| chr11_-_102826434 | 0.58 |
ENST00000340273.4
ENST00000260302.3 |
MMP13
|
matrix metallopeptidase 13 (collagenase 3) |
| chr19_+_2819943 | 0.58 |
ENST00000591265.1
|
ZNF554
|
zinc finger protein 554 |
| chr4_-_120225600 | 0.58 |
ENST00000399075.4
|
C4orf3
|
chromosome 4 open reading frame 3 |
| chr5_-_159846399 | 0.58 |
ENST00000297151.4
|
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr1_-_19229248 | 0.57 |
ENST00000375341.3
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr8_+_52730143 | 0.57 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chrX_+_107288280 | 0.56 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr1_+_16083154 | 0.56 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr20_+_58179582 | 0.56 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr2_+_201450591 | 0.56 |
ENST00000374700.2
|
AOX1
|
aldehyde oxidase 1 |
| chr17_-_36358166 | 0.55 |
ENST00000537432.1
|
TBC1D3
|
TBC1 domain family, member 3 |
| chr2_+_170440902 | 0.54 |
ENST00000448752.2
ENST00000418888.1 ENST00000414307.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr1_-_201346761 | 0.54 |
ENST00000455702.1
ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2
|
troponin T type 2 (cardiac) |
| chr2_+_171036635 | 0.54 |
ENST00000484338.2
ENST00000334231.6 |
MYO3B
|
myosin IIIB |
| chr4_-_186696425 | 0.54 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr14_-_60636561 | 0.54 |
ENST00000536410.2
ENST00000216500.5 |
DHRS7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr2_+_170440844 | 0.53 |
ENST00000260970.3
ENST00000433207.1 ENST00000409714.3 ENST00000462903.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr15_-_82939157 | 0.53 |
ENST00000559949.1
|
RP13-996F3.5
|
golgin A6 family-like 18 |
| chr6_+_116691001 | 0.53 |
ENST00000537543.1
|
DSE
|
dermatan sulfate epimerase |
| chr2_+_113816685 | 0.52 |
ENST00000393200.2
|
IL36RN
|
interleukin 36 receptor antagonist |
| chrM_+_10464 | 0.50 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr19_-_58220517 | 0.50 |
ENST00000512439.2
ENST00000426889.1 |
ZNF154
|
zinc finger protein 154 |
| chr4_+_75174204 | 0.50 |
ENST00000332112.4
ENST00000514968.1 ENST00000503098.1 ENST00000502358.1 ENST00000509145.1 ENST00000505212.1 |
EPGN
|
epithelial mitogen |
| chr12_+_106751436 | 0.49 |
ENST00000228347.4
|
POLR3B
|
polymerase (RNA) III (DNA directed) polypeptide B |
| chr18_+_21032781 | 0.49 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr18_-_53257027 | 0.49 |
ENST00000568740.1
ENST00000564403.2 ENST00000537578.1 |
TCF4
|
transcription factor 4 |
| chr5_-_13944652 | 0.49 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr4_-_186696561 | 0.49 |
ENST00000445115.1
ENST00000451701.1 ENST00000457247.1 ENST00000435480.1 ENST00000425679.1 ENST00000457934.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr20_+_30102231 | 0.48 |
ENST00000335574.5
ENST00000340852.5 ENST00000398174.3 ENST00000376127.3 ENST00000344042.5 |
HM13
|
histocompatibility (minor) 13 |
| chr3_-_128902729 | 0.48 |
ENST00000451728.2
ENST00000446936.2 ENST00000502976.1 ENST00000500450.2 ENST00000441626.2 |
CNBP
|
CCHC-type zinc finger, nucleic acid binding protein |
| chr4_-_47983519 | 0.48 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr19_-_49016418 | 0.47 |
ENST00000270238.3
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr2_+_105050794 | 0.47 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chrM_+_5824 | 0.47 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr12_+_19358228 | 0.47 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr19_+_2819854 | 0.47 |
ENST00000317243.5
|
ZNF554
|
zinc finger protein 554 |
| chr3_+_171561127 | 0.46 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr20_+_12989822 | 0.46 |
ENST00000378194.4
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr15_-_39486510 | 0.46 |
ENST00000560743.1
|
RP11-265N7.1
|
RP11-265N7.1 |
| chr21_+_42742429 | 0.45 |
ENST00000418103.1
|
MX2
|
myxovirus (influenza virus) resistance 2 (mouse) |
| chr1_-_19229014 | 0.45 |
ENST00000538839.1
ENST00000290597.5 |
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr14_-_74417096 | 0.45 |
ENST00000286544.3
|
FAM161B
|
family with sequence similarity 161, member B |
| chr6_-_128841503 | 0.45 |
ENST00000368215.3
ENST00000532331.1 ENST00000368213.5 ENST00000368207.3 ENST00000525459.1 ENST00000368210.3 ENST00000368226.4 ENST00000368227.3 |
PTPRK
|
protein tyrosine phosphatase, receptor type, K |
| chr22_-_50683381 | 0.45 |
ENST00000439308.2
|
TUBGCP6
|
tubulin, gamma complex associated protein 6 |
| chr3_+_577893 | 0.44 |
ENST00000420823.1
ENST00000442809.1 |
AC090044.1
|
AC090044.1 |
| chr1_-_24194771 | 0.44 |
ENST00000374479.3
|
FUCA1
|
fucosidase, alpha-L- 1, tissue |
| chr11_-_26593677 | 0.44 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr5_+_81601166 | 0.43 |
ENST00000439350.1
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr14_-_36990061 | 0.43 |
ENST00000546983.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr21_-_16254231 | 0.42 |
ENST00000412426.1
ENST00000418954.1 |
AF127936.7
|
AF127936.7 |
| chr4_-_74853897 | 0.42 |
ENST00000296028.3
|
PPBP
|
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr1_+_1846519 | 0.42 |
ENST00000378604.3
|
CALML6
|
calmodulin-like 6 |
| chr19_+_44455368 | 0.42 |
ENST00000591168.1
ENST00000587682.1 ENST00000251269.5 |
ZNF221
|
zinc finger protein 221 |
| chr20_+_56964253 | 0.42 |
ENST00000395802.3
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr1_-_43424500 | 0.42 |
ENST00000415851.2
ENST00000426263.3 ENST00000372500.3 |
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1 |
| chr21_-_27945562 | 0.42 |
ENST00000299340.4
ENST00000435845.2 |
CYYR1
|
cysteine/tyrosine-rich 1 |
| chr7_+_44646177 | 0.42 |
ENST00000443864.2
ENST00000447398.1 ENST00000449767.1 ENST00000419661.1 |
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr15_+_42697018 | 0.41 |
ENST00000397204.4
|
CAPN3
|
calpain 3, (p94) |
| chr3_+_182983126 | 0.41 |
ENST00000481531.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr3_+_138068051 | 0.41 |
ENST00000474559.1
|
MRAS
|
muscle RAS oncogene homolog |
| chr5_+_140588269 | 0.41 |
ENST00000541609.1
ENST00000239450.2 |
PCDHB12
|
protocadherin beta 12 |
| chr9_-_3469181 | 0.41 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr21_+_30968360 | 0.40 |
ENST00000333765.4
|
GRIK1-AS2
|
GRIK1 antisense RNA 2 |
| chr2_+_173724771 | 0.40 |
ENST00000538974.1
ENST00000540783.1 |
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr8_+_94767072 | 0.40 |
ENST00000452276.1
ENST00000453321.3 ENST00000498673.1 ENST00000518319.1 |
TMEM67
|
transmembrane protein 67 |
| chr4_-_140223614 | 0.40 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr1_-_27998689 | 0.40 |
ENST00000339145.4
ENST00000362020.4 ENST00000361157.6 |
IFI6
|
interferon, alpha-inducible protein 6 |
| chr10_-_75118611 | 0.40 |
ENST00000355577.3
ENST00000394865.1 ENST00000310715.3 ENST00000401621.2 |
TTC18
|
tetratricopeptide repeat domain 18 |
| chr4_-_76957214 | 0.40 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr12_+_56435637 | 0.40 |
ENST00000356464.5
ENST00000552361.1 |
RPS26
|
ribosomal protein S26 |
| chr16_+_4784458 | 0.39 |
ENST00000590191.1
|
C16orf71
|
chromosome 16 open reading frame 71 |
| chr17_+_34171081 | 0.39 |
ENST00000585577.1
|
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr1_-_24306768 | 0.39 |
ENST00000374453.3
ENST00000453840.3 |
SRSF10
|
serine/arginine-rich splicing factor 10 |
| chr3_-_71294304 | 0.39 |
ENST00000498215.1
|
FOXP1
|
forkhead box P1 |
| chr3_+_53528659 | 0.38 |
ENST00000350061.5
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr5_-_41794313 | 0.38 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr11_+_71791359 | 0.38 |
ENST00000419228.1
ENST00000435085.1 ENST00000307198.7 ENST00000538413.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr7_+_44646162 | 0.38 |
ENST00000439616.2
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr10_-_4285923 | 0.38 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr3_+_101818088 | 0.38 |
ENST00000491959.1
|
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr7_+_102988082 | 0.38 |
ENST00000292644.3
ENST00000544811.1 |
PSMC2
|
proteasome (prosome, macropain) 26S subunit, ATPase, 2 |
| chr8_-_37411648 | 0.37 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chrX_-_9734004 | 0.37 |
ENST00000467482.1
ENST00000380929.2 |
GPR143
|
G protein-coupled receptor 143 |
| chr3_-_156840776 | 0.37 |
ENST00000471357.1
|
LINC00880
|
long intergenic non-protein coding RNA 880 |
| chr4_-_82393052 | 0.37 |
ENST00000335927.7
ENST00000504863.1 ENST00000264400.2 |
RASGEF1B
|
RasGEF domain family, member 1B |
| chr11_-_26593779 | 0.37 |
ENST00000529533.1
|
MUC15
|
mucin 15, cell surface associated |
| chr17_+_66521936 | 0.37 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr1_+_145883868 | 0.37 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chr4_-_153332886 | 0.37 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr2_-_151344172 | 0.37 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr16_+_22501658 | 0.37 |
ENST00000415833.2
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr1_+_149239529 | 0.37 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr2_+_231090433 | 0.36 |
ENST00000486687.2
ENST00000350136.5 ENST00000392045.3 ENST00000417495.3 ENST00000343805.6 ENST00000420434.3 |
SP140
|
SP140 nuclear body protein |
| chr4_+_146403912 | 0.36 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr16_-_15474904 | 0.36 |
ENST00000534094.1
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr5_-_115872124 | 0.36 |
ENST00000515009.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr6_+_37321823 | 0.36 |
ENST00000487950.1
ENST00000469731.1 |
RNF8
|
ring finger protein 8, E3 ubiquitin protein ligase |
| chr19_+_4198072 | 0.35 |
ENST00000262970.5
|
ANKRD24
|
ankyrin repeat domain 24 |
| chr19_-_57988871 | 0.35 |
ENST00000596831.1
ENST00000601768.1 ENST00000356584.3 ENST00000600175.1 ENST00000425074.3 ENST00000343280.4 ENST00000427512.2 |
AC004076.9
ZNF772
|
Uncharacterized protein zinc finger protein 772 |
| chr1_-_178840157 | 0.35 |
ENST00000367629.1
ENST00000234816.2 |
ANGPTL1
|
angiopoietin-like 1 |
| chr6_+_44355257 | 0.35 |
ENST00000371477.3
|
CDC5L
|
cell division cycle 5-like |
| chr17_+_8339164 | 0.34 |
ENST00000582665.1
ENST00000334527.7 ENST00000299734.7 |
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXQ1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.2 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 1.3 | 3.9 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 1.2 | 4.8 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 1.0 | 3.0 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of auditory receptor cell differentiation(GO:0045608) regulation of timing of neuron differentiation(GO:0060164) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.9 | 2.7 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.4 | 2.2 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.4 | 1.3 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.3 | 1.0 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.3 | 1.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.3 | 6.4 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.3 | 0.8 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.3 | 0.8 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.2 | 1.4 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.2 | 0.8 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.2 | 1.4 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.2 | 0.9 | GO:1903284 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 0.5 | GO:0045957 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 0.2 | 0.7 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.2 | 0.5 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.2 | 1.0 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.2 | 0.9 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 0.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.7 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.1 | 0.6 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 0.5 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 1.9 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 0.8 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 0.9 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.5 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.6 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.5 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 1.7 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.3 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 1.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.4 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 1.7 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 1.5 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 1.0 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 0.8 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 1.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.3 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.1 | 0.4 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 0.9 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.1 | 1.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.2 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.1 | 1.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.3 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.5 | GO:1902714 | antifungal humoral response(GO:0019732) negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.1 | 0.4 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.4 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.1 | 1.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.4 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 0.5 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.4 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.1 | 0.3 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 0.4 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.6 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.3 | GO:0006311 | meiotic gene conversion(GO:0006311) male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 0.3 | GO:0070343 | white fat cell proliferation(GO:0070343) positive regulation of fat cell proliferation(GO:0070346) regulation of white fat cell proliferation(GO:0070350) |
| 0.1 | 0.3 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.1 | 0.3 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.3 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.4 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 2.3 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.0 | 0.1 | GO:1903217 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.0 | 0.4 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.7 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 1.1 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 3.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0043132 | NAD transport(GO:0043132) |
| 0.0 | 0.3 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.2 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.2 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 2.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.3 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.7 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.6 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 1.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.7 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.3 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.2 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.6 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0015881 | creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 0.0 | 0.2 | GO:0072093 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 0.4 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.4 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.6 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.2 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.2 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.2 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.6 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.1 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.1 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.3 | GO:0007632 | visual behavior(GO:0007632) |
| 0.0 | 0.2 | GO:0071105 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.0 | 0.8 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.7 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 2.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.6 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.3 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.4 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.0 | 3.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 1.4 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.0 | 0.9 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) oxygen metabolic process(GO:0072592) |
| 0.0 | 0.6 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.1 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 1.1 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 1.0 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.2 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 0.5 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.0 | 1.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.3 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.2 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 1.9 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 4.0 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.3 | GO:2000111 | regulation of macrophage apoptotic process(GO:2000109) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.4 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.2 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.3 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.1 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.0 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 1.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.6 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.4 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.8 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.2 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.0 | 0.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 1.0 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.0 | 0.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.2 | 4.4 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 1.7 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 2.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 1.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.5 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.1 | 0.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.1 | GO:0070470 | plasma membrane respiratory chain(GO:0070470) |
| 0.1 | 1.0 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.3 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.3 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.1 | 0.2 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 0.8 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 0.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 4.0 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.4 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 1.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.4 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 7.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.4 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.0 | 0.3 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 2.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.6 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 1.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 1.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 6.1 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.0 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.8 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.4 | 2.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.4 | 1.4 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.3 | 1.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.3 | 1.0 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.3 | 1.5 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.3 | 1.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.3 | 1.3 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.3 | 0.8 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.2 | 0.7 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.2 | 0.7 | GO:0035643 | L-DOPA receptor activity(GO:0035643) L-DOPA binding(GO:0072544) |
| 0.2 | 1.7 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 1.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.2 | 0.9 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 0.5 | GO:0004877 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.2 | 4.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.4 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 1.0 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.6 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 0.8 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.6 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 0.6 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.5 | GO:0008184 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.5 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 0.8 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 0.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.4 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.4 | GO:0016295 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.1 | 0.3 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 0.3 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 0.4 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 1.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.4 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 1.0 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.5 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 1.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.3 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.1 | 6.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.5 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 0.8 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 1.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 1.0 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 1.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.2 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 1.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.3 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 1.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 3.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.8 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.4 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.4 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 1.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 1.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 6.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.8 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 1.0 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 1.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 1.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 2.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.5 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.8 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0047946 | glutamine N-acyltransferase activity(GO:0047946) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 5.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.4 | GO:0055103 | ligase regulator activity(GO:0055103) |
| 0.0 | 0.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 4.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 2.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 1.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.3 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.0 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 2.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 3.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 5.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 12.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 2.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 2.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 2.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 4.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 4.8 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 2.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.8 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 2.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.8 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.6 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 3.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 1.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 1.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.1 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 1.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |