Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
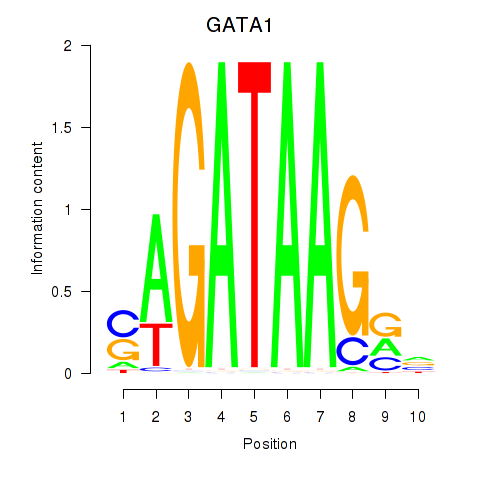
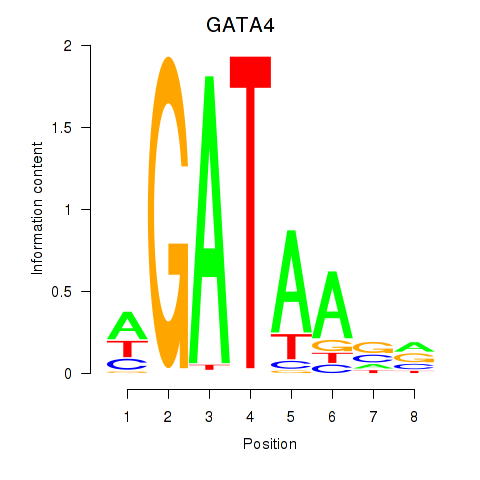
Results for GATA1_GATA4
Z-value: 0.85
Transcription factors associated with GATA1_GATA4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA1
|
ENSG00000102145.9 | GATA binding protein 1 |
|
GATA4
|
ENSG00000136574.13 | GATA binding protein 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GATA4 | hg19_v2_chr8_+_11534462_11534475 | -0.84 | 4.1e-06 | Click! |
| GATA1 | hg19_v2_chrX_+_48644962_48644983 | 0.05 | 8.4e-01 | Click! |
Activity profile of GATA1_GATA4 motif
Sorted Z-values of GATA1_GATA4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_124194752 | 4.49 |
ENST00000318462.6
|
FAM83A
|
family with sequence similarity 83, member A |
| chr1_-_209975494 | 4.29 |
ENST00000456314.1
|
IRF6
|
interferon regulatory factor 6 |
| chr8_+_124194875 | 4.25 |
ENST00000522648.1
ENST00000276699.6 |
FAM83A
|
family with sequence similarity 83, member A |
| chr10_+_71075516 | 4.19 |
ENST00000436817.1
|
HK1
|
hexokinase 1 |
| chr1_-_201368653 | 3.82 |
ENST00000367313.3
|
LAD1
|
ladinin 1 |
| chr1_-_201368707 | 3.75 |
ENST00000391967.2
|
LAD1
|
ladinin 1 |
| chr14_-_81893734 | 3.06 |
ENST00000555447.1
|
STON2
|
stonin 2 |
| chr11_+_34642656 | 2.73 |
ENST00000257831.3
ENST00000450654.2 |
EHF
|
ets homologous factor |
| chr2_+_192543694 | 2.46 |
ENST00000435931.1
|
NABP1
|
nucleic acid binding protein 1 |
| chrX_+_152082969 | 2.39 |
ENST00000535861.1
ENST00000539731.1 ENST00000449285.2 ENST00000318504.7 ENST00000324823.6 ENST00000370268.4 ENST00000370270.2 |
ZNF185
|
zinc finger protein 185 (LIM domain) |
| chr10_+_71075552 | 2.22 |
ENST00000298649.3
|
HK1
|
hexokinase 1 |
| chr12_+_4385230 | 2.12 |
ENST00000536537.1
|
CCND2
|
cyclin D2 |
| chr7_-_87936195 | 2.04 |
ENST00000414498.1
ENST00000301959.5 ENST00000380079.4 |
STEAP4
|
STEAP family member 4 |
| chr6_-_31846744 | 2.03 |
ENST00000414427.1
ENST00000229729.6 ENST00000375562.4 |
SLC44A4
|
solute carrier family 44, member 4 |
| chr1_+_209602156 | 2.01 |
ENST00000429156.1
ENST00000366437.3 ENST00000603283.1 ENST00000431096.1 |
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr5_+_147443534 | 1.99 |
ENST00000398454.1
ENST00000359874.3 ENST00000508733.1 ENST00000256084.7 |
SPINK5
|
serine peptidase inhibitor, Kazal type 5 |
| chr12_-_52845910 | 1.97 |
ENST00000252252.3
|
KRT6B
|
keratin 6B |
| chr1_+_35247859 | 1.93 |
ENST00000373362.3
|
GJB3
|
gap junction protein, beta 3, 31kDa |
| chr19_-_51487071 | 1.88 |
ENST00000391807.1
ENST00000593904.1 |
KLK7
|
kallikrein-related peptidase 7 |
| chr19_-_51522955 | 1.86 |
ENST00000358789.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr1_-_153113927 | 1.77 |
ENST00000368752.4
|
SPRR2B
|
small proline-rich protein 2B |
| chr17_-_79817091 | 1.77 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr1_-_226187013 | 1.74 |
ENST00000272091.7
|
SDE2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr12_-_15103621 | 1.66 |
ENST00000536592.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr11_-_119993979 | 1.56 |
ENST00000524816.3
ENST00000525327.1 |
TRIM29
|
tripartite motif containing 29 |
| chr4_-_68749745 | 1.52 |
ENST00000283916.6
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr19_-_51523275 | 1.51 |
ENST00000309958.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr1_-_175162048 | 1.50 |
ENST00000444639.1
|
KIAA0040
|
KIAA0040 |
| chrX_+_56258844 | 1.48 |
ENST00000374928.3
|
KLF8
|
Kruppel-like factor 8 |
| chr3_+_189507460 | 1.46 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr17_+_74381343 | 1.46 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr19_-_36822551 | 1.43 |
ENST00000591372.1
|
LINC00665
|
long intergenic non-protein coding RNA 665 |
| chr19_-_51487282 | 1.43 |
ENST00000595820.1
ENST00000597707.1 ENST00000336317.4 |
KLK7
|
kallikrein-related peptidase 7 |
| chr15_+_49715449 | 1.42 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr3_-_196987309 | 1.41 |
ENST00000453607.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr2_+_102928009 | 1.41 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr6_+_33168189 | 1.38 |
ENST00000444757.1
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr10_+_5566916 | 1.38 |
ENST00000315238.1
|
CALML3
|
calmodulin-like 3 |
| chr3_+_189507523 | 1.38 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr4_-_159094194 | 1.37 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr11_+_12302492 | 1.34 |
ENST00000533534.1
|
MICALCL
|
MICAL C-terminal like |
| chr6_+_27356745 | 1.33 |
ENST00000461521.1
|
ZNF391
|
zinc finger protein 391 |
| chr10_+_28822417 | 1.33 |
ENST00000428935.1
ENST00000420266.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr14_+_85996507 | 1.29 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr14_+_21510385 | 1.29 |
ENST00000298690.4
|
RNASE7
|
ribonuclease, RNase A family, 7 |
| chr1_-_209825674 | 1.29 |
ENST00000367030.3
ENST00000356082.4 |
LAMB3
|
laminin, beta 3 |
| chr7_-_20256965 | 1.27 |
ENST00000400331.5
ENST00000332878.4 |
MACC1
|
metastasis associated in colon cancer 1 |
| chr10_+_28822636 | 1.26 |
ENST00000442148.1
ENST00000448193.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr5_+_170210721 | 1.25 |
ENST00000265294.4
ENST00000519385.1 ENST00000519598.1 |
GABRP
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr22_-_31688381 | 1.24 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr3_-_69249863 | 1.24 |
ENST00000478263.1
ENST00000462512.1 |
FRMD4B
|
FERM domain containing 4B |
| chr5_+_125695805 | 1.21 |
ENST00000513040.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr14_+_85996471 | 1.18 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr8_+_107630340 | 1.18 |
ENST00000497705.1
|
OXR1
|
oxidation resistance 1 |
| chr7_-_143892748 | 1.17 |
ENST00000378115.2
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35 |
| chr1_-_109849612 | 1.16 |
ENST00000357155.1
|
MYBPHL
|
myosin binding protein H-like |
| chr11_+_34654011 | 1.16 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr4_-_68749699 | 1.13 |
ENST00000545541.1
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr2_+_102927962 | 1.12 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr10_+_180643 | 1.12 |
ENST00000509513.2
ENST00000397959.3 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr1_-_153029980 | 1.12 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr2_+_235903480 | 1.11 |
ENST00000454947.1
|
SH3BP4
|
SH3-domain binding protein 4 |
| chr4_-_77996032 | 1.10 |
ENST00000505609.1
|
CCNI
|
cyclin I |
| chr11_-_119993734 | 1.09 |
ENST00000533302.1
|
TRIM29
|
tripartite motif containing 29 |
| chr4_+_169552748 | 1.07 |
ENST00000504519.1
ENST00000512127.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr5_-_73937244 | 1.07 |
ENST00000302351.4
ENST00000510316.1 ENST00000508331.1 |
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr22_-_29784519 | 1.05 |
ENST00000357586.2
ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr1_-_153013588 | 1.05 |
ENST00000360379.3
|
SPRR2D
|
small proline-rich protein 2D |
| chr3_+_189507432 | 1.05 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chrX_-_78622805 | 1.04 |
ENST00000373298.2
|
ITM2A
|
integral membrane protein 2A |
| chr1_+_158979792 | 1.04 |
ENST00000359709.3
ENST00000430894.2 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr9_-_21974820 | 1.04 |
ENST00000579122.1
ENST00000498124.1 |
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr7_+_144052381 | 1.03 |
ENST00000498580.1
ENST00000056217.5 |
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5 |
| chr12_+_56477093 | 1.03 |
ENST00000549672.1
ENST00000415288.2 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr12_+_6419877 | 1.01 |
ENST00000536531.1
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr15_+_41136216 | 1.00 |
ENST00000562057.1
ENST00000344051.4 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr8_-_128231299 | 0.99 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr1_+_158979680 | 0.98 |
ENST00000368131.4
ENST00000340979.6 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr1_+_158979686 | 0.97 |
ENST00000368132.3
ENST00000295809.7 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr5_+_49961727 | 0.95 |
ENST00000505697.2
ENST00000503750.2 ENST00000514342.2 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr18_+_21452804 | 0.94 |
ENST00000269217.6
|
LAMA3
|
laminin, alpha 3 |
| chrX_+_99899180 | 0.94 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr22_-_31688431 | 0.92 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr2_+_192543153 | 0.92 |
ENST00000425611.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr9_+_132099158 | 0.90 |
ENST00000444125.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr18_+_61445205 | 0.90 |
ENST00000431370.1
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr1_+_209859510 | 0.89 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr3_-_168864427 | 0.89 |
ENST00000468789.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr21_-_34915084 | 0.88 |
ENST00000426819.1
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr4_+_154387480 | 0.88 |
ENST00000409663.3
ENST00000440693.1 ENST00000409959.3 |
KIAA0922
|
KIAA0922 |
| chr19_-_36822595 | 0.87 |
ENST00000585356.1
ENST00000438368.2 ENST00000590622.1 |
LINC00665
|
long intergenic non-protein coding RNA 665 |
| chr7_-_3083573 | 0.85 |
ENST00000396946.4
|
CARD11
|
caspase recruitment domain family, member 11 |
| chr11_+_5712234 | 0.84 |
ENST00000414641.1
|
TRIM22
|
tripartite motif containing 22 |
| chr22_-_43042968 | 0.84 |
ENST00000407623.3
ENST00000396303.3 ENST00000438270.1 |
CYB5R3
|
cytochrome b5 reductase 3 |
| chr17_+_75449889 | 0.82 |
ENST00000590938.1
|
SEPT9
|
septin 9 |
| chr22_-_43042955 | 0.82 |
ENST00000402438.1
|
CYB5R3
|
cytochrome b5 reductase 3 |
| chr14_+_24584056 | 0.80 |
ENST00000561001.1
|
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr1_-_94050668 | 0.80 |
ENST00000539242.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr1_+_174846570 | 0.80 |
ENST00000392064.2
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr15_+_59279851 | 0.79 |
ENST00000348370.4
ENST00000434298.1 ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr19_-_51568324 | 0.78 |
ENST00000595547.1
ENST00000335422.3 ENST00000595793.1 ENST00000596955.1 |
KLK13
|
kallikrein-related peptidase 13 |
| chr1_-_116383738 | 0.77 |
ENST00000320238.3
|
NHLH2
|
nescient helix loop helix 2 |
| chr15_-_53002007 | 0.77 |
ENST00000561490.1
|
FAM214A
|
family with sequence similarity 214, member A |
| chr6_-_112194484 | 0.77 |
ENST00000518295.1
ENST00000484067.2 ENST00000229470.5 ENST00000356013.2 ENST00000368678.4 ENST00000523238.1 ENST00000354650.3 |
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr1_+_81771806 | 0.77 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr6_-_47010061 | 0.76 |
ENST00000371253.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr15_-_34610962 | 0.75 |
ENST00000290209.5
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr10_+_28822236 | 0.75 |
ENST00000347934.4
ENST00000354911.4 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr9_+_71819927 | 0.74 |
ENST00000535702.1
|
TJP2
|
tight junction protein 2 |
| chr3_-_176914191 | 0.74 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr15_-_64126084 | 0.74 |
ENST00000560316.1
ENST00000443617.2 ENST00000560462.1 ENST00000558532.1 ENST00000561400.1 |
HERC1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr5_+_131409476 | 0.73 |
ENST00000296871.2
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr18_+_61445007 | 0.72 |
ENST00000447428.1
ENST00000546027.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr14_+_24584372 | 0.72 |
ENST00000559396.1
ENST00000558638.1 ENST00000561041.1 ENST00000559288.1 ENST00000558408.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chrX_-_24665353 | 0.72 |
ENST00000379144.2
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr4_+_160188306 | 0.72 |
ENST00000510510.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr7_-_3083472 | 0.72 |
ENST00000356408.3
|
CARD11
|
caspase recruitment domain family, member 11 |
| chr2_-_45236540 | 0.71 |
ENST00000303077.6
|
SIX2
|
SIX homeobox 2 |
| chr6_+_130339710 | 0.71 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr5_+_50679506 | 0.70 |
ENST00000511384.1
|
ISL1
|
ISL LIM homeobox 1 |
| chr1_-_116926718 | 0.70 |
ENST00000598661.1
|
AL136376.1
|
Uncharacterized protein |
| chr1_+_25599018 | 0.70 |
ENST00000417538.2
ENST00000357542.4 ENST00000568195.1 ENST00000342055.5 ENST00000423810.2 |
RHD
|
Rh blood group, D antigen |
| chr4_+_123073464 | 0.69 |
ENST00000264501.4
|
KIAA1109
|
KIAA1109 |
| chr6_-_11779014 | 0.69 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr12_+_104359641 | 0.69 |
ENST00000537100.1
|
TDG
|
thymine-DNA glycosylase |
| chr4_+_79475019 | 0.68 |
ENST00000508214.1
|
ANXA3
|
annexin A3 |
| chr11_-_57102947 | 0.68 |
ENST00000526696.1
|
SSRP1
|
structure specific recognition protein 1 |
| chr12_-_67197760 | 0.68 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr4_-_143226979 | 0.67 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr4_-_143227088 | 0.67 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr21_+_39628852 | 0.67 |
ENST00000398938.2
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr12_+_104337515 | 0.66 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr11_+_34663913 | 0.66 |
ENST00000532302.1
|
EHF
|
ets homologous factor |
| chr12_-_122879969 | 0.65 |
ENST00000540304.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr15_-_79237433 | 0.65 |
ENST00000220166.5
|
CTSH
|
cathepsin H |
| chr8_-_72268721 | 0.65 |
ENST00000419131.1
ENST00000388743.2 |
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr1_-_214638146 | 0.65 |
ENST00000543945.1
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr20_+_51588873 | 0.65 |
ENST00000371497.5
|
TSHZ2
|
teashirt zinc finger homeobox 2 |
| chr15_-_64665911 | 0.65 |
ENST00000606793.1
ENST00000561349.1 ENST00000560278.1 |
CTD-2116N17.1
|
Uncharacterized protein |
| chr17_-_40288449 | 0.64 |
ENST00000552162.1
ENST00000550504.1 |
RAB5C
|
RAB5C, member RAS oncogene family |
| chr11_+_10477733 | 0.64 |
ENST00000528723.1
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr5_-_39219641 | 0.63 |
ENST00000509072.1
ENST00000504542.1 ENST00000505428.1 ENST00000506557.1 |
FYB
|
FYN binding protein |
| chr2_+_201242715 | 0.63 |
ENST00000421573.1
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr11_-_26588634 | 0.63 |
ENST00000436318.2
ENST00000281268.8 |
MUC15
|
mucin 15, cell surface associated |
| chr6_-_11779403 | 0.63 |
ENST00000414691.3
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr6_-_47009996 | 0.63 |
ENST00000371243.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr6_+_43112037 | 0.62 |
ENST00000473339.1
|
PTK7
|
protein tyrosine kinase 7 |
| chr5_-_39270725 | 0.62 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr7_+_128379346 | 0.62 |
ENST00000535011.2
ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU
|
calumenin |
| chr10_+_63808970 | 0.61 |
ENST00000309334.5
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr1_-_163172625 | 0.61 |
ENST00000527988.1
ENST00000531476.1 ENST00000530507.1 |
RGS5
|
regulator of G-protein signaling 5 |
| chr14_-_85996332 | 0.61 |
ENST00000380722.1
|
RP11-497E19.1
|
RP11-497E19.1 |
| chr12_+_4430371 | 0.61 |
ENST00000179259.4
|
C12orf5
|
chromosome 12 open reading frame 5 |
| chr11_-_105892937 | 0.61 |
ENST00000301919.4
ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr11_+_64685026 | 0.60 |
ENST00000526559.1
|
PPP2R5B
|
protein phosphatase 2, regulatory subunit B', beta |
| chr2_+_170440902 | 0.60 |
ENST00000448752.2
ENST00000418888.1 ENST00000414307.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr4_+_25657444 | 0.60 |
ENST00000504570.1
ENST00000382051.3 |
SLC34A2
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 2 |
| chr19_-_41934635 | 0.60 |
ENST00000321702.2
|
B3GNT8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
| chr12_-_95010147 | 0.59 |
ENST00000548918.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr6_-_111927449 | 0.59 |
ENST00000368761.5
ENST00000392556.4 ENST00000340026.6 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr7_+_123469037 | 0.59 |
ENST00000489978.1
|
HYAL4
|
hyaluronoglucosaminidase 4 |
| chr4_-_83821676 | 0.58 |
ENST00000355196.2
ENST00000507676.1 ENST00000506495.1 ENST00000507051.1 |
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr11_+_128563652 | 0.57 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr1_+_19967014 | 0.57 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr11_+_128563948 | 0.57 |
ENST00000534087.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr15_-_72462705 | 0.56 |
ENST00000564129.1
|
GRAMD2
|
GRAM domain containing 2 |
| chr5_+_67576062 | 0.55 |
ENST00000523807.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr13_+_53029564 | 0.55 |
ENST00000468284.1
ENST00000378034.3 ENST00000258607.5 ENST00000378037.5 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr4_+_74702214 | 0.55 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr19_+_37096194 | 0.55 |
ENST00000460670.1
ENST00000292928.2 ENST00000439428.1 |
ZNF382
|
zinc finger protein 382 |
| chr16_+_14280564 | 0.54 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr6_+_30614886 | 0.54 |
ENST00000376471.4
|
C6orf136
|
chromosome 6 open reading frame 136 |
| chr13_-_107214291 | 0.54 |
ENST00000375926.1
|
ARGLU1
|
arginine and glutamate rich 1 |
| chr12_+_25205155 | 0.54 |
ENST00000550945.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chrX_-_24665208 | 0.54 |
ENST00000356768.4
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr1_-_152009460 | 0.53 |
ENST00000271638.2
|
S100A11
|
S100 calcium binding protein A11 |
| chr15_+_64752927 | 0.52 |
ENST00000416172.1
|
ZNF609
|
zinc finger protein 609 |
| chr15_-_72462661 | 0.52 |
ENST00000570275.1
|
GRAMD2
|
GRAM domain containing 2 |
| chr19_-_58485895 | 0.52 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr6_+_30614779 | 0.51 |
ENST00000293604.6
ENST00000376473.5 |
C6orf136
|
chromosome 6 open reading frame 136 |
| chr8_-_29605625 | 0.51 |
ENST00000506121.3
|
LINC00589
|
long intergenic non-protein coding RNA 589 |
| chr10_+_180405 | 0.51 |
ENST00000439456.1
ENST00000397962.3 ENST00000309776.4 ENST00000381602.4 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr4_+_75174180 | 0.50 |
ENST00000413830.1
|
EPGN
|
epithelial mitogen |
| chr12_+_104359614 | 0.50 |
ENST00000266775.9
ENST00000544861.1 |
TDG
|
thymine-DNA glycosylase |
| chr2_+_170440844 | 0.50 |
ENST00000260970.3
ENST00000433207.1 ENST00000409714.3 ENST00000462903.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr6_-_150039170 | 0.49 |
ENST00000458696.2
ENST00000392273.3 |
LATS1
|
large tumor suppressor kinase 1 |
| chr1_-_161337662 | 0.49 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr2_+_234600253 | 0.49 |
ENST00000373424.1
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr2_-_214016314 | 0.49 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr12_-_30907862 | 0.49 |
ENST00000541765.1
ENST00000537108.1 |
CAPRIN2
|
caprin family member 2 |
| chr10_+_114710516 | 0.49 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr19_+_35739897 | 0.49 |
ENST00000605618.1
ENST00000427250.1 ENST00000601623.1 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr21_-_34915123 | 0.49 |
ENST00000438059.1
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr1_-_204116078 | 0.48 |
ENST00000367198.2
ENST00000452983.1 |
ETNK2
|
ethanolamine kinase 2 |
| chr9_+_33795533 | 0.48 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr6_+_7108210 | 0.48 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr6_-_170599561 | 0.48 |
ENST00000366756.3
|
DLL1
|
delta-like 1 (Drosophila) |
| chr7_-_27169801 | 0.48 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr15_-_99057551 | 0.48 |
ENST00000558256.1
|
FAM169B
|
family with sequence similarity 169, member B |
| chr1_+_31883048 | 0.48 |
ENST00000536859.1
|
SERINC2
|
serine incorporator 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA1_GATA4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.8 | 3.3 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.8 | 5.3 | GO:0002786 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.6 | 6.6 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.5 | 1.5 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.4 | 2.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.4 | 2.5 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.4 | 2.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.4 | 1.5 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.4 | 2.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.3 | 1.6 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.3 | 1.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.3 | 1.1 | GO:0048936 | visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) cardiac cell fate determination(GO:0060913) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.3 | 2.0 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.2 | 0.7 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.2 | 0.7 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.2 | 1.7 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.2 | 1.4 | GO:0060437 | lung growth(GO:0060437) |
| 0.2 | 1.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.2 | 0.7 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.2 | 1.3 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.2 | 0.8 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.2 | 1.0 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.2 | 0.7 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.2 | 0.5 | GO:0048633 | negative regulation of auditory receptor cell differentiation(GO:0045608) positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.2 | 2.2 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 0.9 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 1.7 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.4 | GO:2000742 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 | 1.4 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 0.6 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.1 | 0.5 | GO:0051413 | response to cortisone(GO:0051413) |
| 0.1 | 0.4 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 | 0.6 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 4.2 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.1 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.6 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.5 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.1 | 0.2 | GO:0007493 | endodermal cell fate determination(GO:0007493) |
| 0.1 | 0.6 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.1 | 1.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 1.6 | GO:0070970 | interleukin-2 secretion(GO:0070970) |
| 0.1 | 0.8 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.1 | 0.7 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 0.8 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.3 | GO:1904000 | positive regulation of growth rate(GO:0040010) positive regulation of eating behavior(GO:1904000) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.1 | 0.7 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.4 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 2.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 0.6 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 0.6 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.3 | GO:1904875 | regulation of DNA ligase activity(GO:1904875) |
| 0.1 | 0.8 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 1.0 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.1 | 0.3 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 0.4 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 0.5 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.4 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 1.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 1.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 1.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.4 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.2 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.1 | 2.4 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.7 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 1.7 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.2 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 0.1 | 0.3 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.1 | 0.7 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.2 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.1 | 0.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 2.0 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 0.6 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.4 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.1 | 2.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.2 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 0.3 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 0.6 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.1 | 0.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.2 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.1 | 0.4 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.8 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 2.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.2 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 0.7 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.5 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 3.1 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 0.1 | 0.7 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 0.2 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.1 | GO:1902613 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.0 | 0.4 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.8 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 1.1 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.4 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.0 | 1.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 1.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.5 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.9 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.3 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.0 | 2.7 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.4 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.5 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.2 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.4 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.4 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.1 | GO:1902769 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.7 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 1.1 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.4 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 2.8 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.4 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.2 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 3.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.9 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 1.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.2 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 1.1 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.1 | GO:0070602 | regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 9.2 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.5 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.4 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.6 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 1.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 1.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 1.3 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.5 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.3 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.2 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 1.1 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 3.4 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.2 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.4 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.0 | 0.2 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.2 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.7 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 1.0 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.4 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 1.0 | GO:0043090 | amino acid import(GO:0043090) |
| 0.0 | 1.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.2 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.4 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.3 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.2 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.2 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.7 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.4 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.9 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.4 | GO:0043086 | negative regulation of catalytic activity(GO:0043086) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.5 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.3 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.1 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.2 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.9 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.0 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.1 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 1.6 | GO:0007049 | cell cycle(GO:0007049) |
| 0.0 | 0.1 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 1.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.0 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.7 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.0 | 0.1 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.0 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.2 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.2 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.2 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.3 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.1 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.0 | 0.1 | GO:0072221 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) |
| 0.0 | 0.8 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.6 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 1.1 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 1.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 1.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.5 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.0 | 0.3 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.5 | 1.9 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.4 | 3.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.3 | 2.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 2.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 6.8 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 1.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 1.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 1.6 | GO:0012505 | endomembrane system(GO:0012505) |
| 0.1 | 0.8 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 1.4 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 1.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.4 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 0.9 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 1.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.7 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.7 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 0.2 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.3 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.1 | 0.3 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 3.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.9 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 1.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 2.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 5.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.7 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.8 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 3.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 2.2 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 1.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 1.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.0 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 2.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 3.3 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.7 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.0 | GO:0036025 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 1.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0031672 | A band(GO:0031672) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.7 | 2.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.7 | 6.4 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.5 | 1.5 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.5 | 9.9 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.4 | 1.7 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.3 | 0.9 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.3 | 2.0 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.3 | 1.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.3 | 2.0 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.2 | 3.9 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 1.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.2 | 1.5 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 1.9 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.2 | 0.8 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 1.7 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.7 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.4 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.1 | 3.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.7 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.3 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 1.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.6 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.6 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 0.3 | GO:0031768 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.1 | 1.9 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.7 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 0.6 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 1.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.8 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.3 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.1 | 0.3 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 3.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.3 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 3.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 1.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.2 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.1 | 0.3 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.1 | 0.7 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 1.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 1.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.6 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 0.5 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 0.5 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.6 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.2 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.1 | 0.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 1.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.6 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.4 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 1.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.5 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.5 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.4 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.9 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.3 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 5.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 1.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.2 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 9.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:0070016 | gamma-catenin binding(GO:0045295) armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.5 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0047756 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.4 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 1.7 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 1.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 3.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.0 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 12.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.4 | GO:0070008 | serine-type carboxypeptidase activity(GO:0004185) serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 1.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.4 | GO:0005261 | cation channel activity(GO:0005261) |
| 0.0 | 3.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.4 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.5 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.4 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 1.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 3.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.8 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 2.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 2.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 2.0 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 3.0 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.9 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.1 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.4 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.5 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 1.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 1.1 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 1.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.9 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 3.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 2.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.7 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.7 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 2.0 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.2 | REACTOME CELL CYCLE CHECKPOINTS | Genes involved in Cell Cycle Checkpoints |
| 0.0 | 1.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |