Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
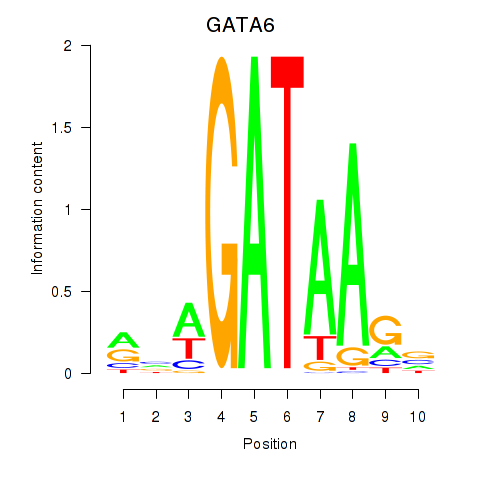
Results for GATA6
Z-value: 0.63
Transcription factors associated with GATA6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA6
|
ENSG00000141448.4 | GATA binding protein 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GATA6 | hg19_v2_chr18_+_19750894_19750980 | -0.56 | 1.1e-02 | Click! |
Activity profile of GATA6 motif
Sorted Z-values of GATA6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_105952648 | 2.70 |
ENST00000330233.7
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr6_+_12290586 | 2.12 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr7_-_107880508 | 2.05 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
| chr9_+_118950325 | 1.79 |
ENST00000534838.1
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr20_+_43029911 | 1.72 |
ENST00000443598.2
ENST00000316099.4 ENST00000415691.2 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr14_+_100204027 | 1.58 |
ENST00000554479.1
ENST00000555145.1 |
EML1
|
echinoderm microtubule associated protein like 1 |
| chr11_+_118958689 | 1.43 |
ENST00000535253.1
ENST00000392841.1 |
HMBS
|
hydroxymethylbilane synthase |
| chr14_-_23285069 | 1.34 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr7_-_100239132 | 1.19 |
ENST00000223051.3
ENST00000431692.1 |
TFR2
|
transferrin receptor 2 |
| chr14_-_23285011 | 1.14 |
ENST00000397532.3
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr8_-_95449155 | 1.11 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr10_-_99531709 | 1.09 |
ENST00000266066.3
|
SFRP5
|
secreted frizzled-related protein 5 |
| chrX_-_10851762 | 1.04 |
ENST00000380785.1
ENST00000380787.1 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr14_-_23284703 | 0.87 |
ENST00000555911.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr17_+_4675175 | 0.85 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr11_-_66206260 | 0.85 |
ENST00000329819.4
ENST00000310999.7 ENST00000430466.2 |
MRPL11
|
mitochondrial ribosomal protein L11 |
| chr18_-_2982869 | 0.84 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr17_+_38673270 | 0.82 |
ENST00000578280.1
|
RP5-1028K7.2
|
RP5-1028K7.2 |
| chr19_-_11494975 | 0.78 |
ENST00000222139.6
ENST00000592375.2 |
EPOR
|
erythropoietin receptor |
| chr11_-_10920838 | 0.78 |
ENST00000503469.2
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr5_+_135496675 | 0.74 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr20_-_54967187 | 0.72 |
ENST00000422322.1
ENST00000371356.2 ENST00000451915.1 ENST00000347343.2 ENST00000395911.1 ENST00000395907.1 ENST00000441357.1 ENST00000456249.1 ENST00000420474.1 ENST00000395909.4 ENST00000395914.1 ENST00000312783.6 ENST00000395915.3 ENST00000395913.3 |
AURKA
|
aurora kinase A |
| chr16_+_616995 | 0.64 |
ENST00000293874.2
ENST00000409527.2 ENST00000424439.2 ENST00000540585.1 |
PIGQ
NHLRC4
|
phosphatidylinositol glycan anchor biosynthesis, class Q NHL repeat containing 4 |
| chr6_-_86099898 | 0.60 |
ENST00000455071.1
|
RP11-30P6.6
|
RP11-30P6.6 |
| chr22_-_20231207 | 0.60 |
ENST00000425986.1
|
RTN4R
|
reticulon 4 receptor |
| chr4_-_103266219 | 0.59 |
ENST00000394833.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr3_+_69812877 | 0.58 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr19_+_36132631 | 0.55 |
ENST00000379026.2
ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2
|
ets variant 2 |
| chr7_+_155090271 | 0.53 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr2_+_109237717 | 0.52 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr17_+_19314505 | 0.50 |
ENST00000461366.1
|
RNF112
|
ring finger protein 112 |
| chr11_-_10920714 | 0.50 |
ENST00000533941.1
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr1_-_24469602 | 0.50 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1 |
| chr1_+_161136180 | 0.49 |
ENST00000352210.5
ENST00000367999.4 ENST00000544598.1 ENST00000535223.1 ENST00000432542.2 |
PPOX
|
protoporphyrinogen oxidase |
| chr1_-_205912577 | 0.49 |
ENST00000367135.3
ENST00000367134.2 |
SLC26A9
|
solute carrier family 26 (anion exchanger), member 9 |
| chr2_+_3642545 | 0.46 |
ENST00000382062.2
ENST00000236693.7 ENST00000349077.4 |
COLEC11
|
collectin sub-family member 11 |
| chr17_+_43224684 | 0.44 |
ENST00000332499.2
|
HEXIM1
|
hexamethylene bis-acetamide inducible 1 |
| chr17_+_56833184 | 0.43 |
ENST00000308249.2
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr4_-_103266355 | 0.42 |
ENST00000424970.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr10_-_127505167 | 0.42 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr5_+_159848807 | 0.41 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chr2_-_219850277 | 0.40 |
ENST00000295727.1
|
FEV
|
FEV (ETS oncogene family) |
| chr8_+_21823726 | 0.40 |
ENST00000433566.4
|
XPO7
|
exportin 7 |
| chr18_-_28742813 | 0.38 |
ENST00000257197.3
ENST00000257198.5 |
DSC1
|
desmocollin 1 |
| chr12_+_122880045 | 0.34 |
ENST00000539034.1
ENST00000535976.1 |
RP11-450K4.1
|
RP11-450K4.1 |
| chr2_-_202298268 | 0.34 |
ENST00000440597.1
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr7_-_142659388 | 0.33 |
ENST00000476829.1
ENST00000467543.1 |
KEL
|
Kell blood group, metallo-endopeptidase |
| chr11_-_47374246 | 0.31 |
ENST00000545968.1
ENST00000399249.2 ENST00000256993.4 |
MYBPC3
|
myosin binding protein C, cardiac |
| chr14_+_53173890 | 0.30 |
ENST00000445930.2
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr17_+_4835580 | 0.29 |
ENST00000329125.5
|
GP1BA
|
glycoprotein Ib (platelet), alpha polypeptide |
| chr19_-_39303576 | 0.29 |
ENST00000594209.1
|
LGALS4
|
lectin, galactoside-binding, soluble, 4 |
| chr14_-_23284675 | 0.26 |
ENST00000555959.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr14_-_38036271 | 0.25 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr17_+_19314432 | 0.24 |
ENST00000575165.2
|
RNF112
|
ring finger protein 112 |
| chr15_+_49715293 | 0.24 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr6_-_46048116 | 0.24 |
ENST00000185206.6
|
CLIC5
|
chloride intracellular channel 5 |
| chr12_-_54689532 | 0.23 |
ENST00000540264.2
ENST00000312156.4 |
NFE2
|
nuclear factor, erythroid 2 |
| chr4_+_15376165 | 0.23 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr16_+_4475806 | 0.23 |
ENST00000262375.6
ENST00000355296.4 ENST00000431375.2 ENST00000574895.1 |
DNAJA3
|
DnaJ (Hsp40) homolog, subfamily A, member 3 |
| chr9_-_114937676 | 0.22 |
ENST00000374270.3
|
SUSD1
|
sushi domain containing 1 |
| chr10_-_21186144 | 0.22 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr17_+_72920698 | 0.22 |
ENST00000580223.1
|
OTOP2
|
otopetrin 2 |
| chr13_+_24144509 | 0.22 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr14_+_50999744 | 0.21 |
ENST00000441560.2
|
ATL1
|
atlastin GTPase 1 |
| chr14_-_21945057 | 0.21 |
ENST00000397762.1
|
RAB2B
|
RAB2B, member RAS oncogene family |
| chr17_-_62084241 | 0.21 |
ENST00000449662.2
|
ICAM2
|
intercellular adhesion molecule 2 |
| chr17_-_38938786 | 0.21 |
ENST00000301656.3
|
KRT27
|
keratin 27 |
| chr2_-_40739501 | 0.20 |
ENST00000403092.1
ENST00000402441.1 ENST00000448531.1 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr16_+_202686 | 0.20 |
ENST00000252951.2
|
HBZ
|
hemoglobin, zeta |
| chr20_-_60294804 | 0.20 |
ENST00000317652.1
|
RP11-429E11.3
|
Uncharacterized protein |
| chr1_+_66999799 | 0.20 |
ENST00000371035.3
ENST00000371036.3 ENST00000371037.4 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr15_-_38852251 | 0.19 |
ENST00000558432.1
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr1_-_167487808 | 0.17 |
ENST00000392122.3
|
CD247
|
CD247 molecule |
| chr1_-_167487758 | 0.17 |
ENST00000362089.5
|
CD247
|
CD247 molecule |
| chr13_+_24144796 | 0.16 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr18_+_43304092 | 0.16 |
ENST00000321925.4
ENST00000587601.1 |
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
| chr2_-_44065889 | 0.16 |
ENST00000543989.1
ENST00000405322.1 |
ABCG5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr5_+_131892603 | 0.15 |
ENST00000378823.3
ENST00000265335.6 |
RAD50
|
RAD50 homolog (S. cerevisiae) |
| chr17_+_72920370 | 0.15 |
ENST00000331427.4
|
OTOP2
|
otopetrin 2 |
| chr8_-_93107443 | 0.15 |
ENST00000360348.2
ENST00000520428.1 ENST00000518992.1 ENST00000520556.1 ENST00000518317.1 ENST00000521319.1 ENST00000521375.1 ENST00000518449.1 |
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr6_-_30128657 | 0.14 |
ENST00000449742.2
ENST00000376704.3 |
TRIM10
|
tripartite motif containing 10 |
| chr4_+_166794383 | 0.14 |
ENST00000061240.2
ENST00000507499.1 |
TLL1
|
tolloid-like 1 |
| chr1_-_42630389 | 0.14 |
ENST00000357001.2
|
GUCA2A
|
guanylate cyclase activator 2A (guanylin) |
| chr2_+_220436917 | 0.14 |
ENST00000243786.2
|
INHA
|
inhibin, alpha |
| chr1_+_171217677 | 0.14 |
ENST00000402921.2
|
FMO1
|
flavin containing monooxygenase 1 |
| chr5_-_135290651 | 0.13 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr22_+_35462129 | 0.13 |
ENST00000404699.2
ENST00000308700.6 |
ISX
|
intestine-specific homeobox |
| chr7_+_142749417 | 0.13 |
ENST00000418316.1
|
OR6V1
|
olfactory receptor, family 6, subfamily V, member 1 |
| chr11_+_10326918 | 0.12 |
ENST00000528544.1
|
ADM
|
adrenomedullin |
| chr15_+_81391740 | 0.11 |
ENST00000561216.1
|
C15orf26
|
chromosome 15 open reading frame 26 |
| chr16_+_71560023 | 0.11 |
ENST00000572450.1
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr9_-_114937543 | 0.11 |
ENST00000374264.2
ENST00000374263.3 |
SUSD1
|
sushi domain containing 1 |
| chr3_+_88188254 | 0.10 |
ENST00000309495.5
|
ZNF654
|
zinc finger protein 654 |
| chr19_-_55549624 | 0.10 |
ENST00000417454.1
ENST00000310373.3 ENST00000333884.2 |
GP6
|
glycoprotein VI (platelet) |
| chr1_+_171217622 | 0.09 |
ENST00000433267.1
ENST00000367750.3 |
FMO1
|
flavin containing monooxygenase 1 |
| chr5_+_131892815 | 0.08 |
ENST00000453394.1
|
RAD50
|
RAD50 homolog (S. cerevisiae) |
| chr16_+_333152 | 0.08 |
ENST00000219406.6
ENST00000404312.1 ENST00000456379.1 |
PDIA2
|
protein disulfide isomerase family A, member 2 |
| chr9_-_117692697 | 0.08 |
ENST00000223795.2
|
TNFSF8
|
tumor necrosis factor (ligand) superfamily, member 8 |
| chr11_+_10326612 | 0.07 |
ENST00000534464.1
ENST00000530439.1 ENST00000524948.1 ENST00000528655.1 ENST00000526492.1 ENST00000525063.1 |
ADM
|
adrenomedullin |
| chr8_-_93107696 | 0.07 |
ENST00000436581.2
ENST00000520583.1 ENST00000519061.1 |
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr1_+_206317591 | 0.07 |
ENST00000432969.2
|
CTSE
|
cathepsin E |
| chr1_-_158656488 | 0.07 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr10_+_118305435 | 0.07 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr7_+_150690857 | 0.07 |
ENST00000484524.1
ENST00000467517.1 |
NOS3
|
nitric oxide synthase 3 (endothelial cell) |
| chr20_+_43935474 | 0.06 |
ENST00000372743.1
ENST00000372741.3 ENST00000343694.3 |
RBPJL
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr16_+_71560154 | 0.06 |
ENST00000539698.3
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr3_-_122694215 | 0.06 |
ENST00000449546.1
|
SEMA5B
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B |
| chr3_+_69812701 | 0.06 |
ENST00000472437.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr16_+_67022476 | 0.06 |
ENST00000540947.2
|
CES4A
|
carboxylesterase 4A |
| chr12_-_6233828 | 0.04 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr2_+_173792893 | 0.04 |
ENST00000535187.1
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr18_+_43304137 | 0.04 |
ENST00000502059.2
ENST00000586951.1 ENST00000589322.2 ENST00000415427.3 ENST00000535474.1 ENST00000402943.2 |
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
| chr5_+_157602404 | 0.04 |
ENST00000522975.1
|
CTC-436K13.1
|
CTC-436K13.1 |
| chr9_-_114937465 | 0.04 |
ENST00000355396.3
|
SUSD1
|
sushi domain containing 1 |
| chr1_-_120354079 | 0.04 |
ENST00000354219.1
ENST00000369401.4 ENST00000256585.5 |
REG4
|
regenerating islet-derived family, member 4 |
| chr12_-_120765565 | 0.04 |
ENST00000423423.3
ENST00000308366.4 |
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr3_+_137717571 | 0.03 |
ENST00000343735.4
|
CLDN18
|
claudin 18 |
| chr11_-_116708302 | 0.01 |
ENST00000375320.1
ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1
|
apolipoprotein A-I |
| chr5_-_135290705 | 0.01 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr2_-_152830441 | 0.01 |
ENST00000534999.1
ENST00000397327.2 |
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr2_-_152830479 | 0.01 |
ENST00000360283.6
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr1_-_27240455 | 0.00 |
ENST00000254227.3
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.5 | 2.1 | GO:0060585 | nitric oxide transport(GO:0030185) negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.4 | 3.6 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.3 | 0.8 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.2 | 0.7 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.2 | 1.2 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.2 | 0.6 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.2 | 0.5 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 1.7 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.2 | 2.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.2 | 1.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 | 2.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.7 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 0.3 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.1 | 0.3 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.1 | 1.0 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.3 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.1 | 0.1 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.1 | 1.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 1.6 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.3 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.6 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.2 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.2 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.6 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 1.6 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.9 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.7 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) |
| 0.0 | 0.2 | GO:0071340 | activation-induced cell death of T cells(GO:0006924) skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.4 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.4 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.5 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.2 | 0.7 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 1.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.3 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 2.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.6 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.7 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.5 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.3 | 1.2 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.2 | 0.6 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.2 | 3.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.4 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 2.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.5 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.7 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 2.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.7 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.5 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 1.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.2 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.2 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 1.7 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 2.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 2.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 2.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.4 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |