Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
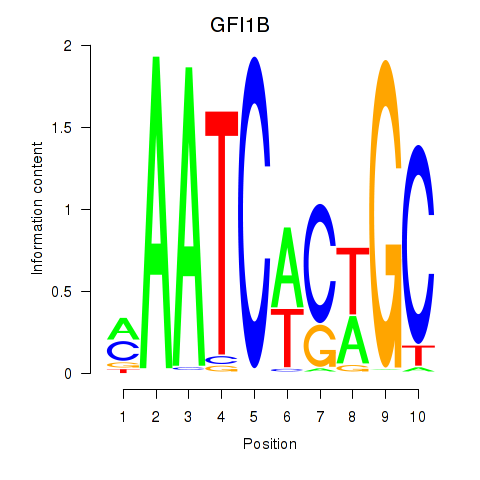
Results for GFI1B
Z-value: 0.56
Transcription factors associated with GFI1B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GFI1B
|
ENSG00000165702.8 | growth factor independent 1B transcriptional repressor |
Activity profile of GFI1B motif
Sorted Z-values of GFI1B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_115301235 | 0.90 |
ENST00000525878.1
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr1_-_220219775 | 0.78 |
ENST00000609181.1
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr11_+_125461607 | 0.76 |
ENST00000527606.1
|
STT3A
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr5_+_109025067 | 0.73 |
ENST00000261483.4
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1 |
| chr19_+_7445850 | 0.68 |
ENST00000593531.1
|
CTD-2207O23.3
|
Rho guanine nucleotide exchange factor 18 |
| chr17_+_46131843 | 0.68 |
ENST00000577411.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr10_-_129924611 | 0.68 |
ENST00000368654.3
|
MKI67
|
marker of proliferation Ki-67 |
| chr12_+_6603253 | 0.67 |
ENST00000382457.4
ENST00000545962.1 |
NCAPD2
|
non-SMC condensin I complex, subunit D2 |
| chr7_+_134576317 | 0.67 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr5_+_150591678 | 0.67 |
ENST00000523466.1
|
GM2A
|
GM2 ganglioside activator |
| chr10_-_129924468 | 0.63 |
ENST00000368653.3
|
MKI67
|
marker of proliferation Ki-67 |
| chr13_+_76362974 | 0.60 |
ENST00000497947.2
|
LMO7
|
LIM domain 7 |
| chr3_-_149688971 | 0.60 |
ENST00000498307.1
ENST00000489155.1 |
PFN2
|
profilin 2 |
| chr3_+_152017924 | 0.59 |
ENST00000465907.2
ENST00000492948.1 ENST00000485509.1 ENST00000464596.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr18_+_56532100 | 0.58 |
ENST00000588456.1
ENST00000589481.1 ENST00000591049.1 |
ZNF532
|
zinc finger protein 532 |
| chr8_+_21823726 | 0.57 |
ENST00000433566.4
|
XPO7
|
exportin 7 |
| chr9_-_19065082 | 0.57 |
ENST00000415524.1
|
HAUS6
|
HAUS augmin-like complex, subunit 6 |
| chr7_+_55086703 | 0.56 |
ENST00000455089.1
ENST00000342916.3 ENST00000344576.2 ENST00000420316.2 |
EGFR
|
epidermal growth factor receptor |
| chr9_-_95055923 | 0.55 |
ENST00000430417.1
|
IARS
|
isoleucyl-tRNA synthetase |
| chr2_+_29341037 | 0.55 |
ENST00000449202.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr5_-_64920115 | 0.54 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr15_-_90456156 | 0.54 |
ENST00000357484.5
|
C15orf38
|
chromosome 15 open reading frame 38 |
| chr2_-_96874553 | 0.53 |
ENST00000337288.5
ENST00000443962.1 |
STARD7
|
StAR-related lipid transfer (START) domain containing 7 |
| chr9_-_95056010 | 0.51 |
ENST00000443024.2
|
IARS
|
isoleucyl-tRNA synthetase |
| chr17_+_46131912 | 0.51 |
ENST00000584634.1
ENST00000580050.1 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr15_-_72514866 | 0.51 |
ENST00000562997.1
|
PKM
|
pyruvate kinase, muscle |
| chr6_-_56707943 | 0.51 |
ENST00000370769.4
ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST
|
dystonin |
| chr1_+_203651937 | 0.49 |
ENST00000341360.2
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr6_-_131384373 | 0.48 |
ENST00000392427.3
ENST00000525271.1 ENST00000527411.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr17_+_46132037 | 0.47 |
ENST00000582155.1
ENST00000583378.1 ENST00000536222.1 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr2_-_110371412 | 0.47 |
ENST00000415095.1
ENST00000334001.6 ENST00000437928.1 ENST00000493445.1 ENST00000397714.2 ENST00000461295.1 |
SEPT10
|
septin 10 |
| chr13_-_67802549 | 0.47 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr7_-_108097144 | 0.47 |
ENST00000418239.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr15_-_59041954 | 0.47 |
ENST00000439637.1
ENST00000558004.1 |
ADAM10
|
ADAM metallopeptidase domain 10 |
| chr16_+_21623392 | 0.46 |
ENST00000562961.1
|
METTL9
|
methyltransferase like 9 |
| chr4_-_909521 | 0.45 |
ENST00000511229.1
|
GAK
|
cyclin G associated kinase |
| chr2_-_112614424 | 0.45 |
ENST00000427997.1
|
ANAPC1
|
anaphase promoting complex subunit 1 |
| chr17_-_5321549 | 0.44 |
ENST00000572809.1
|
NUP88
|
nucleoporin 88kDa |
| chr2_-_208030295 | 0.44 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr17_+_48133330 | 0.43 |
ENST00000544892.1
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr2_-_208030886 | 0.43 |
ENST00000426163.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr14_-_53417732 | 0.43 |
ENST00000399304.3
ENST00000395631.2 ENST00000341590.3 ENST00000343279.4 |
FERMT2
|
fermitin family member 2 |
| chr2_+_9563769 | 0.43 |
ENST00000475482.1
|
CPSF3
|
cleavage and polyadenylation specific factor 3, 73kDa |
| chr17_+_44803922 | 0.41 |
ENST00000465370.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr10_+_82219058 | 0.41 |
ENST00000372156.1
|
TSPAN14
|
tetraspanin 14 |
| chr1_-_179834311 | 0.41 |
ENST00000553856.1
|
IFRG15
|
Homo sapiens torsin A interacting protein 2 (TOR1AIP2), transcript variant 1, mRNA. |
| chr6_-_131384412 | 0.41 |
ENST00000445890.2
ENST00000368128.2 ENST00000337057.3 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr10_+_17272608 | 0.40 |
ENST00000421459.2
|
VIM
|
vimentin |
| chr10_-_91403625 | 0.39 |
ENST00000322191.6
ENST00000342512.3 ENST00000371774.2 |
PANK1
|
pantothenate kinase 1 |
| chr1_-_94703118 | 0.39 |
ENST00000260526.6
ENST00000370217.3 |
ARHGAP29
|
Rho GTPase activating protein 29 |
| chr6_+_32937083 | 0.39 |
ENST00000456339.1
|
BRD2
|
bromodomain containing 2 |
| chr6_-_131384347 | 0.38 |
ENST00000530481.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr1_+_35544968 | 0.38 |
ENST00000359858.4
ENST00000373330.1 |
ZMYM1
|
zinc finger, MYM-type 1 |
| chr6_-_133119649 | 0.38 |
ENST00000367918.1
|
SLC18B1
|
solute carrier family 18, subfamily B, member 1 |
| chr1_+_164529004 | 0.37 |
ENST00000559240.1
ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr15_-_70994612 | 0.37 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr12_+_100967420 | 0.37 |
ENST00000266754.5
ENST00000547754.1 |
GAS2L3
|
growth arrest-specific 2 like 3 |
| chr1_+_244816371 | 0.37 |
ENST00000263831.7
|
DESI2
|
desumoylating isopeptidase 2 |
| chr11_+_93861993 | 0.36 |
ENST00000227638.3
ENST00000436171.2 |
PANX1
|
pannexin 1 |
| chr12_-_57006882 | 0.36 |
ENST00000551996.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr3_-_33759699 | 0.36 |
ENST00000399362.4
ENST00000359576.5 ENST00000307312.7 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr14_-_103434869 | 0.36 |
ENST00000559043.1
|
CDC42BPB
|
CDC42 binding protein kinase beta (DMPK-like) |
| chr1_+_164528616 | 0.36 |
ENST00000340699.3
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr1_-_21620877 | 0.36 |
ENST00000527991.1
|
ECE1
|
endothelin converting enzyme 1 |
| chr11_+_35684288 | 0.35 |
ENST00000299413.5
|
TRIM44
|
tripartite motif containing 44 |
| chr16_+_68279256 | 0.35 |
ENST00000564827.2
ENST00000566188.1 ENST00000444212.2 ENST00000568082.1 |
PLA2G15
|
phospholipase A2, group XV |
| chrX_-_54069576 | 0.35 |
ENST00000445025.1
ENST00000322659.8 |
PHF8
|
PHD finger protein 8 |
| chr15_+_67458861 | 0.34 |
ENST00000558428.1
ENST00000558827.1 |
SMAD3
|
SMAD family member 3 |
| chr7_+_112063192 | 0.34 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr10_+_31610064 | 0.34 |
ENST00000446923.2
ENST00000559476.1 |
ZEB1
|
zinc finger E-box binding homeobox 1 |
| chr4_+_108852697 | 0.33 |
ENST00000508453.1
|
CYP2U1
|
cytochrome P450, family 2, subfamily U, polypeptide 1 |
| chr6_+_32936942 | 0.33 |
ENST00000496118.2
|
BRD2
|
bromodomain containing 2 |
| chr21_+_35014706 | 0.33 |
ENST00000399353.1
ENST00000444491.1 ENST00000381318.3 |
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr14_-_64194745 | 0.33 |
ENST00000247225.6
|
SGPP1
|
sphingosine-1-phosphate phosphatase 1 |
| chr1_+_74663896 | 0.33 |
ENST00000370898.3
ENST00000467578.2 ENST00000370894.5 ENST00000482102.2 ENST00000609362.1 ENST00000534056.1 ENST00000557284.2 ENST00000370899.3 ENST00000370895.1 ENST00000534632.1 ENST00000370893.1 ENST00000370891.2 |
FPGT
FPGT-TNNI3K
TNNI3K
|
fucose-1-phosphate guanylyltransferase FPGT-TNNI3K readthrough TNNI3 interacting kinase |
| chr22_+_19467261 | 0.32 |
ENST00000455750.1
ENST00000437685.2 ENST00000263201.1 ENST00000404724.3 |
CDC45
|
cell division cycle 45 |
| chr7_-_112579673 | 0.32 |
ENST00000432572.1
|
C7orf60
|
chromosome 7 open reading frame 60 |
| chr6_+_107077471 | 0.32 |
ENST00000369044.1
|
QRSL1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr17_+_75450099 | 0.32 |
ENST00000586433.1
|
SEPT9
|
septin 9 |
| chr2_+_54683419 | 0.32 |
ENST00000356805.4
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr1_+_10290822 | 0.32 |
ENST00000377083.1
|
KIF1B
|
kinesin family member 1B |
| chr3_-_149688655 | 0.32 |
ENST00000461930.1
ENST00000423691.2 ENST00000490975.1 ENST00000461868.1 ENST00000452853.2 |
PFN2
|
profilin 2 |
| chr22_+_19466980 | 0.32 |
ENST00000407835.1
ENST00000438587.1 |
CDC45
|
cell division cycle 45 |
| chr11_-_61582579 | 0.31 |
ENST00000539419.1
ENST00000545245.1 ENST00000545405.1 ENST00000542506.1 |
FADS1
|
fatty acid desaturase 1 |
| chr11_-_13461790 | 0.31 |
ENST00000530907.1
|
BTBD10
|
BTB (POZ) domain containing 10 |
| chr18_+_56532053 | 0.31 |
ENST00000592452.1
|
ZNF532
|
zinc finger protein 532 |
| chr16_+_88493879 | 0.31 |
ENST00000565624.1
ENST00000437464.1 |
ZNF469
|
zinc finger protein 469 |
| chr11_+_76571911 | 0.31 |
ENST00000534206.1
ENST00000532485.1 ENST00000526597.1 ENST00000533873.1 ENST00000538157.1 |
ACER3
|
alkaline ceramidase 3 |
| chr17_-_1395954 | 0.31 |
ENST00000359786.5
|
MYO1C
|
myosin IC |
| chr17_+_75449889 | 0.31 |
ENST00000590938.1
|
SEPT9
|
septin 9 |
| chr9_-_123638633 | 0.30 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19 |
| chr7_-_108096765 | 0.30 |
ENST00000379024.4
ENST00000351718.4 |
NRCAM
|
neuronal cell adhesion molecule |
| chr1_+_164528437 | 0.30 |
ENST00000485769.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr17_+_75450075 | 0.29 |
ENST00000592951.1
|
SEPT9
|
septin 9 |
| chr5_+_79703823 | 0.29 |
ENST00000338008.5
ENST00000510158.1 ENST00000505560.1 |
ZFYVE16
|
zinc finger, FYVE domain containing 16 |
| chr9_-_14180778 | 0.29 |
ENST00000380924.1
ENST00000543693.1 |
NFIB
|
nuclear factor I/B |
| chr5_-_60458179 | 0.29 |
ENST00000507416.1
ENST00000339020.3 |
SMIM15
|
small integral membrane protein 15 |
| chr5_-_125930877 | 0.29 |
ENST00000510111.2
ENST00000509270.1 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr17_+_76210367 | 0.29 |
ENST00000592734.1
ENST00000587746.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr3_-_119813264 | 0.29 |
ENST00000264235.8
|
GSK3B
|
glycogen synthase kinase 3 beta |
| chr6_-_131321863 | 0.29 |
ENST00000528282.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr1_-_47134085 | 0.29 |
ENST00000371937.4
ENST00000574428.1 ENST00000329231.4 |
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr17_-_79481666 | 0.28 |
ENST00000575659.1
|
ACTG1
|
actin, gamma 1 |
| chr7_+_134576151 | 0.28 |
ENST00000393118.2
|
CALD1
|
caldesmon 1 |
| chr10_+_94590910 | 0.28 |
ENST00000371547.4
|
EXOC6
|
exocyst complex component 6 |
| chr19_+_36545781 | 0.28 |
ENST00000388999.3
|
WDR62
|
WD repeat domain 62 |
| chr1_-_92764523 | 0.28 |
ENST00000370360.3
ENST00000534881.1 |
GLMN
|
glomulin, FKBP associated protein |
| chr14_-_80677815 | 0.28 |
ENST00000557125.1
ENST00000555750.1 |
DIO2
|
deiodinase, iodothyronine, type II |
| chr12_-_117175819 | 0.28 |
ENST00000261318.3
ENST00000536380.1 |
C12orf49
|
chromosome 12 open reading frame 49 |
| chr1_+_210406121 | 0.27 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4 |
| chr6_-_56708459 | 0.27 |
ENST00000370788.2
|
DST
|
dystonin |
| chr7_-_108096822 | 0.27 |
ENST00000379028.3
ENST00000413765.2 ENST00000379022.4 |
NRCAM
|
neuronal cell adhesion molecule |
| chr2_-_190044480 | 0.27 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr18_+_11857439 | 0.27 |
ENST00000602628.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr1_+_150337144 | 0.26 |
ENST00000539519.1
ENST00000369067.3 ENST00000369068.4 |
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr2_-_110371720 | 0.26 |
ENST00000356688.4
|
SEPT10
|
septin 10 |
| chr15_+_71228826 | 0.26 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr4_+_26165074 | 0.26 |
ENST00000512351.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr12_-_21910775 | 0.26 |
ENST00000539782.1
|
LDHB
|
lactate dehydrogenase B |
| chr7_+_79765071 | 0.26 |
ENST00000457358.2
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr2_-_208030647 | 0.26 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr14_-_68162438 | 0.26 |
ENST00000557273.1
ENST00000428130.2 |
RDH11
|
retinol dehydrogenase 11 (all-trans/9-cis/11-cis) |
| chr11_-_33774944 | 0.26 |
ENST00000532057.1
ENST00000531080.1 |
FBXO3
|
F-box protein 3 |
| chr13_-_27745936 | 0.25 |
ENST00000282344.6
|
USP12
|
ubiquitin specific peptidase 12 |
| chr1_+_33283246 | 0.25 |
ENST00000526230.1
ENST00000531256.1 ENST00000482212.1 |
S100PBP
|
S100P binding protein |
| chr1_-_197169672 | 0.25 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chrX_-_71933888 | 0.25 |
ENST00000373542.4
ENST00000339490.3 ENST00000541944.1 ENST00000373539.3 ENST00000373545.3 |
PHKA1
|
phosphorylase kinase, alpha 1 (muscle) |
| chr2_-_110371664 | 0.25 |
ENST00000545389.1
ENST00000423520.1 |
SEPT10
|
septin 10 |
| chr7_+_150758642 | 0.24 |
ENST00000488420.1
|
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr11_+_74699703 | 0.24 |
ENST00000529024.1
ENST00000544263.1 |
NEU3
|
sialidase 3 (membrane sialidase) |
| chr8_-_28965684 | 0.24 |
ENST00000523130.1
|
KIF13B
|
kinesin family member 13B |
| chrX_+_57618269 | 0.24 |
ENST00000374888.1
|
ZXDB
|
zinc finger, X-linked, duplicated B |
| chr12_+_32638897 | 0.24 |
ENST00000531134.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr2_+_47630108 | 0.23 |
ENST00000233146.2
ENST00000454849.1 ENST00000543555.1 |
MSH2
|
mutS homolog 2 |
| chr6_-_116601044 | 0.23 |
ENST00000368608.3
|
TSPYL1
|
TSPY-like 1 |
| chr7_+_66093908 | 0.23 |
ENST00000443322.1
ENST00000449064.1 |
KCTD7
|
potassium channel tetramerization domain containing 7 |
| chr1_-_205091115 | 0.23 |
ENST00000264515.6
ENST00000367164.1 |
RBBP5
|
retinoblastoma binding protein 5 |
| chr2_-_110371777 | 0.23 |
ENST00000397712.2
|
SEPT10
|
septin 10 |
| chr2_-_55496174 | 0.23 |
ENST00000417363.1
ENST00000412530.1 ENST00000394600.3 ENST00000366137.2 ENST00000420637.1 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr15_-_38519066 | 0.23 |
ENST00000561320.1
ENST00000561161.1 |
RP11-346D14.1
|
RP11-346D14.1 |
| chr20_+_34679725 | 0.23 |
ENST00000432589.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr1_-_220220000 | 0.23 |
ENST00000366923.3
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr7_+_55086794 | 0.23 |
ENST00000275493.2
ENST00000442591.1 |
EGFR
|
epidermal growth factor receptor |
| chr17_-_79895097 | 0.23 |
ENST00000402252.2
ENST00000583564.1 ENST00000585244.1 ENST00000337943.5 ENST00000579698.1 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr1_-_74663825 | 0.22 |
ENST00000370911.3
ENST00000370909.2 ENST00000354431.4 |
LRRIQ3
|
leucine-rich repeats and IQ motif containing 3 |
| chr2_-_10978103 | 0.22 |
ENST00000404824.2
|
PDIA6
|
protein disulfide isomerase family A, member 6 |
| chr4_-_102268484 | 0.22 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr8_+_87354945 | 0.22 |
ENST00000517970.1
|
WWP1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr2_+_201994042 | 0.22 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr3_+_50284321 | 0.21 |
ENST00000451956.1
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr14_+_72052983 | 0.21 |
ENST00000358550.2
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr3_+_152017360 | 0.21 |
ENST00000485910.1
ENST00000463374.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr11_+_70116779 | 0.21 |
ENST00000253925.7
ENST00000389547.3 |
PPFIA1
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 |
| chrX_+_46696372 | 0.21 |
ENST00000218340.3
|
RP2
|
retinitis pigmentosa 2 (X-linked recessive) |
| chr2_-_176033066 | 0.21 |
ENST00000437522.1
|
ATF2
|
activating transcription factor 2 |
| chr18_+_55712915 | 0.21 |
ENST00000592846.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr4_+_48807155 | 0.20 |
ENST00000504654.1
|
OCIAD1
|
OCIA domain containing 1 |
| chr18_-_48723690 | 0.20 |
ENST00000406189.3
|
MEX3C
|
mex-3 RNA binding family member C |
| chr14_-_80677970 | 0.20 |
ENST00000438257.4
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr1_+_40810516 | 0.20 |
ENST00000435168.2
|
SMAP2
|
small ArfGAP2 |
| chr6_-_13621126 | 0.20 |
ENST00000600057.1
|
AL441883.1
|
Uncharacterized protein |
| chr3_-_149688896 | 0.20 |
ENST00000239940.7
|
PFN2
|
profilin 2 |
| chr6_-_36842784 | 0.20 |
ENST00000373699.5
|
PPIL1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr10_+_72575643 | 0.19 |
ENST00000373202.3
|
SGPL1
|
sphingosine-1-phosphate lyase 1 |
| chr19_+_57862622 | 0.19 |
ENST00000391705.3
ENST00000443917.2 ENST00000598744.1 |
ZNF304
|
zinc finger protein 304 |
| chr21_+_35014783 | 0.19 |
ENST00000381291.4
ENST00000381285.4 ENST00000399367.3 ENST00000399352.1 ENST00000399355.2 ENST00000399349.1 |
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr17_+_5973740 | 0.19 |
ENST00000576083.1
|
WSCD1
|
WSC domain containing 1 |
| chr12_-_123752624 | 0.19 |
ENST00000542174.1
ENST00000535796.1 |
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1 |
| chr4_+_41937131 | 0.19 |
ENST00000504986.1
ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33
|
transmembrane protein 33 |
| chr15_-_75918787 | 0.19 |
ENST00000564086.1
|
SNUPN
|
snurportin 1 |
| chr1_+_206516200 | 0.19 |
ENST00000295713.5
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr2_-_86116020 | 0.19 |
ENST00000525834.2
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr9_-_123239632 | 0.19 |
ENST00000416449.1
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr15_-_59041768 | 0.19 |
ENST00000402627.1
ENST00000396140.2 ENST00000559053.1 ENST00000561288.1 |
ADAM10
|
ADAM metallopeptidase domain 10 |
| chr12_+_117348742 | 0.18 |
ENST00000309909.5
ENST00000455858.2 |
FBXW8
|
F-box and WD repeat domain containing 8 |
| chr1_-_68698197 | 0.18 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr17_-_79895154 | 0.18 |
ENST00000405481.4
ENST00000585215.1 ENST00000577624.1 ENST00000403172.4 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr17_-_56595196 | 0.18 |
ENST00000579921.1
ENST00000579925.1 ENST00000323456.5 |
MTMR4
|
myotubularin related protein 4 |
| chr17_+_79031415 | 0.18 |
ENST00000572073.1
ENST00000573677.1 |
BAIAP2
|
BAI1-associated protein 2 |
| chr19_+_55477711 | 0.18 |
ENST00000448584.2
ENST00000537859.1 ENST00000585500.1 ENST00000427260.2 ENST00000538819.1 ENST00000263437.6 |
NLRP2
|
NLR family, pyrin domain containing 2 |
| chr5_+_98109322 | 0.18 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr9_-_92051391 | 0.18 |
ENST00000420681.2
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr17_-_295730 | 0.18 |
ENST00000329099.4
|
FAM101B
|
family with sequence similarity 101, member B |
| chr14_+_58765305 | 0.18 |
ENST00000445108.1
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr4_+_15471489 | 0.18 |
ENST00000424120.1
ENST00000413206.1 ENST00000438599.2 ENST00000511544.1 ENST00000512702.1 ENST00000507954.1 ENST00000515124.1 ENST00000503292.1 ENST00000503658.1 |
CC2D2A
|
coiled-coil and C2 domain containing 2A |
| chr9_-_104145795 | 0.18 |
ENST00000259407.2
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr12_-_56843161 | 0.18 |
ENST00000554616.1
ENST00000553532.1 ENST00000229201.4 |
TIMELESS
|
timeless circadian clock |
| chr14_-_64108125 | 0.18 |
ENST00000267522.3
|
WDR89
|
WD repeat domain 89 |
| chr15_+_67458357 | 0.17 |
ENST00000537194.2
|
SMAD3
|
SMAD family member 3 |
| chr19_+_58258164 | 0.17 |
ENST00000317178.5
|
ZNF776
|
zinc finger protein 776 |
| chr18_-_44500101 | 0.17 |
ENST00000589917.1
ENST00000587810.1 |
PIAS2
|
protein inhibitor of activated STAT, 2 |
| chr3_+_184055240 | 0.17 |
ENST00000383847.2
|
FAM131A
|
family with sequence similarity 131, member A |
| chr1_+_45792541 | 0.17 |
ENST00000334815.3
|
HPDL
|
4-hydroxyphenylpyruvate dioxygenase-like |
| chr6_-_138820624 | 0.17 |
ENST00000343505.5
|
NHSL1
|
NHS-like 1 |
| chr6_+_21593972 | 0.17 |
ENST00000244745.1
ENST00000543472.1 |
SOX4
|
SRY (sex determining region Y)-box 4 |
| chr10_+_88516396 | 0.17 |
ENST00000372037.3
|
BMPR1A
|
bone morphogenetic protein receptor, type IA |
| chr4_+_41362796 | 0.17 |
ENST00000508501.1
ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr17_+_19281787 | 0.17 |
ENST00000482850.1
|
MAPK7
|
mitogen-activated protein kinase 7 |
| chr3_+_159481464 | 0.17 |
ENST00000467377.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr2_+_86116396 | 0.16 |
ENST00000455121.3
|
AC105053.4
|
AC105053.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GFI1B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.3 | 1.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.2 | 0.6 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.2 | 0.8 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.2 | 1.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 0.5 | GO:2000283 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.2 | 0.8 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.4 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.7 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 | 0.5 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.1 | 0.3 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 0.4 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.4 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.3 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 0.3 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.4 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.3 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 | 0.3 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.1 | 0.9 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 1.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.5 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.2 | GO:1902559 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.1 | 1.7 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.3 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 0.3 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.3 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.1 | 1.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.2 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 | 0.4 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.4 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.2 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.1 | 0.5 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.7 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.2 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.1 | 0.3 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.1 | 0.3 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.2 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) |
| 0.1 | 0.3 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.1 | 0.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.8 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.2 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.4 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.2 | GO:0048378 | lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) |
| 0.0 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.2 | GO:0061015 | snRNA import into nucleus(GO:0061015) |
| 0.0 | 0.5 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 1.7 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.3 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.6 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.3 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.4 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.3 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 1.0 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0039650 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.3 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.1 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.3 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.9 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.1 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 1.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0090071 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.3 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.4 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.2 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.4 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.5 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.3 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.3 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.0 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.3 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.2 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.1 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.2 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.2 | 0.6 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.2 | 0.7 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.1 | 0.4 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 0.8 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.5 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.1 | 1.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.7 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 1.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 1.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.7 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.4 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.1 | 0.2 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.1 | 1.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 1.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.4 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 0.2 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.1 | 0.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 1.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.8 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.4 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.3 | 1.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.2 | 0.8 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 0.5 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.2 | 0.8 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.7 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.1 | 0.4 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 1.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.3 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.2 | GO:0000406 | double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.1 | 0.6 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.7 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.5 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.4 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.2 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.1 | 0.4 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.2 | GO:0047291 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.0 | 0.3 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.8 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.2 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 1.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.6 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.2 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 1.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 1.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.5 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.3 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.0 | GO:0000994 | RNA polymerase III core binding(GO:0000994) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) all-trans retinal binding(GO:0005503) |
| 0.0 | 0.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.7 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.8 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.9 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |