Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
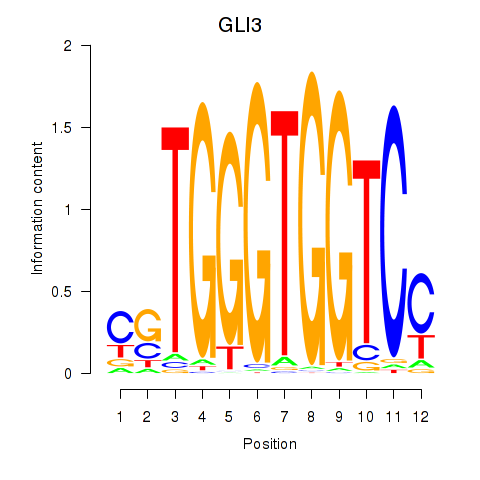
Results for GLI3
Z-value: 2.08
Transcription factors associated with GLI3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLI3
|
ENSG00000106571.8 | GLI family zinc finger 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLI3 | hg19_v2_chr7_-_42276612_42276782 | 0.90 | 5.2e-08 | Click! |
Activity profile of GLI3 motif
Sorted Z-values of GLI3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_153538292 | 12.81 |
ENST00000497140.1
ENST00000368708.3 |
S100A2
|
S100 calcium binding protein A2 |
| chr12_-_52887034 | 12.06 |
ENST00000330722.6
|
KRT6A
|
keratin 6A |
| chr18_+_21529811 | 9.56 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr1_+_183155373 | 8.42 |
ENST00000493293.1
ENST00000264144.4 |
LAMC2
|
laminin, gamma 2 |
| chr1_-_153538011 | 8.39 |
ENST00000368707.4
|
S100A2
|
S100 calcium binding protein A2 |
| chr4_-_90758227 | 8.37 |
ENST00000506691.1
ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr12_-_52867569 | 7.33 |
ENST00000252250.6
|
KRT6C
|
keratin 6C |
| chr19_+_35606692 | 6.73 |
ENST00000406242.3
ENST00000454903.2 |
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chr12_-_52845910 | 6.66 |
ENST00000252252.3
|
KRT6B
|
keratin 6B |
| chr17_-_39928106 | 6.64 |
ENST00000540235.1
|
JUP
|
junction plakoglobin |
| chr16_+_68679193 | 6.44 |
ENST00000581171.1
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental) |
| chr19_-_51456344 | 6.39 |
ENST00000336334.3
ENST00000593428.1 |
KLK5
|
kallikrein-related peptidase 5 |
| chr1_-_1356628 | 6.26 |
ENST00000442470.1
ENST00000537107.1 |
ANKRD65
|
ankyrin repeat domain 65 |
| chr1_+_152881014 | 5.71 |
ENST00000368764.3
ENST00000392667.2 |
IVL
|
involucrin |
| chr20_+_62327996 | 5.51 |
ENST00000369996.1
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy |
| chr19_-_51456321 | 5.32 |
ENST00000391809.2
|
KLK5
|
kallikrein-related peptidase 5 |
| chr16_+_68678739 | 5.20 |
ENST00000264012.4
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental) |
| chr21_-_28215332 | 5.11 |
ENST00000517777.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr11_-_118550375 | 5.06 |
ENST00000525958.1
ENST00000264029.4 ENST00000397925.1 ENST00000529101.1 |
TREH
|
trehalase (brush-border membrane glycoprotein) |
| chr6_-_32784687 | 4.85 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr3_-_195538760 | 4.84 |
ENST00000475231.1
|
MUC4
|
mucin 4, cell surface associated |
| chr11_-_119187826 | 4.79 |
ENST00000264036.4
|
MCAM
|
melanoma cell adhesion molecule |
| chr16_+_55522536 | 4.74 |
ENST00000570283.1
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
| chr20_-_1306351 | 4.59 |
ENST00000381812.1
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr8_+_124194752 | 4.58 |
ENST00000318462.6
|
FAM83A
|
family with sequence similarity 83, member A |
| chrX_-_100914781 | 4.50 |
ENST00000431597.1
ENST00000458024.1 ENST00000413506.1 ENST00000440675.1 ENST00000328766.5 ENST00000356824.4 |
ARMCX2
|
armadillo repeat containing, X-linked 2 |
| chr8_+_124194875 | 4.48 |
ENST00000522648.1
ENST00000276699.6 |
FAM83A
|
family with sequence similarity 83, member A |
| chr16_-_68269971 | 4.32 |
ENST00000565858.1
|
ESRP2
|
epithelial splicing regulatory protein 2 |
| chr3_-_195538728 | 4.21 |
ENST00000349607.4
ENST00000346145.4 |
MUC4
|
mucin 4, cell surface associated |
| chr1_+_13910194 | 4.06 |
ENST00000376057.4
ENST00000510906.1 |
PDPN
|
podoplanin |
| chr12_-_54779511 | 4.04 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr20_-_1306391 | 4.01 |
ENST00000339987.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr13_-_20767037 | 3.98 |
ENST00000382848.4
|
GJB2
|
gap junction protein, beta 2, 26kDa |
| chr19_-_20844368 | 3.96 |
ENST00000595094.1
ENST00000601440.1 ENST00000291750.6 |
CTC-513N18.7
ZNF626
|
CTC-513N18.7 zinc finger protein 626 |
| chr1_-_160990886 | 3.94 |
ENST00000537746.1
|
F11R
|
F11 receptor |
| chr20_+_44098385 | 3.92 |
ENST00000217425.5
ENST00000339946.3 |
WFDC2
|
WAP four-disulfide core domain 2 |
| chr1_-_209792111 | 3.86 |
ENST00000455193.1
|
LAMB3
|
laminin, beta 3 |
| chr5_-_149792295 | 3.85 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr19_+_48281842 | 3.67 |
ENST00000509570.2
|
SEPW1
|
selenoprotein W, 1 |
| chr1_+_36789335 | 3.58 |
ENST00000373137.2
|
RP11-268J15.5
|
RP11-268J15.5 |
| chr20_+_44637526 | 3.55 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr5_+_9546306 | 3.55 |
ENST00000508179.1
|
SNHG18
|
small nucleolar RNA host gene 18 (non-protein coding) |
| chr19_-_53324884 | 3.51 |
ENST00000457749.2
ENST00000414252.2 ENST00000391783.2 |
ZNF28
|
zinc finger protein 28 |
| chr1_+_156123359 | 3.49 |
ENST00000368284.1
ENST00000368286.2 ENST00000438830.1 |
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr16_-_2908155 | 3.45 |
ENST00000571228.1
ENST00000161006.3 |
PRSS22
|
protease, serine, 22 |
| chr19_-_51456198 | 3.42 |
ENST00000594846.1
|
KLK5
|
kallikrein-related peptidase 5 |
| chr1_+_152956549 | 3.42 |
ENST00000307122.2
|
SPRR1A
|
small proline-rich protein 1A |
| chr7_-_41742697 | 3.39 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr12_-_57634475 | 3.34 |
ENST00000393825.1
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 |
| chr19_-_35323762 | 3.33 |
ENST00000590963.1
|
CTC-523E23.4
|
CTC-523E23.4 |
| chrX_+_135618258 | 3.32 |
ENST00000440515.1
ENST00000456412.1 |
VGLL1
|
vestigial like 1 (Drosophila) |
| chr1_-_85358850 | 3.31 |
ENST00000370611.3
|
LPAR3
|
lysophosphatidic acid receptor 3 |
| chr20_+_44098346 | 3.29 |
ENST00000372676.3
|
WFDC2
|
WAP four-disulfide core domain 2 |
| chr11_-_119991589 | 3.27 |
ENST00000526881.1
|
TRIM29
|
tripartite motif containing 29 |
| chr1_-_1356719 | 3.24 |
ENST00000520296.1
|
ANKRD65
|
ankyrin repeat domain 65 |
| chr19_-_11689752 | 3.24 |
ENST00000592659.1
ENST00000592828.1 ENST00000218758.5 ENST00000412435.2 |
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr16_+_2880157 | 3.22 |
ENST00000382280.3
|
ZG16B
|
zymogen granule protein 16B |
| chr1_+_44399347 | 3.21 |
ENST00000477048.1
ENST00000471394.2 |
ARTN
|
artemin |
| chr1_-_201368707 | 3.21 |
ENST00000391967.2
|
LAD1
|
ladinin 1 |
| chr19_-_51472031 | 3.18 |
ENST00000391808.1
|
KLK6
|
kallikrein-related peptidase 6 |
| chr6_-_159420780 | 3.14 |
ENST00000449822.1
|
RSPH3
|
radial spoke 3 homolog (Chlamydomonas) |
| chr21_-_33984456 | 3.12 |
ENST00000431216.1
ENST00000553001.1 ENST00000440966.1 |
AP000275.65
C21orf59
|
Uncharacterized protein chromosome 21 open reading frame 59 |
| chr16_+_68771128 | 3.08 |
ENST00000261769.5
ENST00000422392.2 |
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr8_+_143761874 | 3.08 |
ENST00000301258.4
ENST00000513264.1 |
PSCA
|
prostate stem cell antigen |
| chr15_-_45422056 | 3.08 |
ENST00000267803.4
ENST00000559014.1 ENST00000558851.1 ENST00000559988.1 ENST00000558996.1 ENST00000558422.1 ENST00000559226.1 ENST00000558326.1 ENST00000558377.1 ENST00000559644.1 |
DUOXA1
|
dual oxidase maturation factor 1 |
| chr11_-_111783919 | 3.07 |
ENST00000531198.1
ENST00000533879.1 |
CRYAB
|
crystallin, alpha B |
| chr15_+_45422178 | 3.04 |
ENST00000389037.3
ENST00000558322.1 |
DUOX1
|
dual oxidase 1 |
| chr22_-_38349552 | 3.01 |
ENST00000422191.1
ENST00000249079.2 ENST00000418863.1 ENST00000403305.1 ENST00000403026.1 |
C22orf23
|
chromosome 22 open reading frame 23 |
| chr5_-_95297678 | 2.99 |
ENST00000237853.4
|
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr5_-_180237445 | 2.98 |
ENST00000393340.3
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chrX_+_115567767 | 2.97 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr22_+_38035623 | 2.93 |
ENST00000336738.5
ENST00000442465.2 |
SH3BP1
|
SH3-domain binding protein 1 |
| chr1_+_35220613 | 2.92 |
ENST00000338513.1
|
GJB5
|
gap junction protein, beta 5, 31.1kDa |
| chr7_-_92465868 | 2.92 |
ENST00000424848.2
|
CDK6
|
cyclin-dependent kinase 6 |
| chr6_-_29648887 | 2.90 |
ENST00000376883.1
|
ZFP57
|
ZFP57 zinc finger protein |
| chr7_-_121784285 | 2.89 |
ENST00000417368.2
|
AASS
|
aminoadipate-semialdehyde synthase |
| chr1_+_40942887 | 2.86 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chr19_-_2050852 | 2.84 |
ENST00000541165.1
ENST00000591601.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr12_-_57030096 | 2.84 |
ENST00000549506.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr11_-_65640325 | 2.80 |
ENST00000307998.6
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2 |
| chrX_-_7895755 | 2.78 |
ENST00000444736.1
ENST00000537427.1 ENST00000442940.1 |
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr10_-_27444143 | 2.78 |
ENST00000477432.1
|
YME1L1
|
YME1-like 1 ATPase |
| chr1_+_44399466 | 2.78 |
ENST00000498139.2
ENST00000491846.1 |
ARTN
|
artemin |
| chr10_+_88718314 | 2.76 |
ENST00000348795.4
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1) |
| chr14_-_61748550 | 2.75 |
ENST00000555868.1
|
TMEM30B
|
transmembrane protein 30B |
| chr10_-_105615164 | 2.73 |
ENST00000355946.2
ENST00000369774.4 |
SH3PXD2A
|
SH3 and PX domains 2A |
| chr5_+_148960931 | 2.72 |
ENST00000333677.6
|
ARHGEF37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr5_-_95297534 | 2.72 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr19_-_35992780 | 2.69 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr15_+_45422131 | 2.67 |
ENST00000321429.4
|
DUOX1
|
dual oxidase 1 |
| chr1_-_62785054 | 2.66 |
ENST00000371153.4
|
KANK4
|
KN motif and ankyrin repeat domains 4 |
| chr1_-_31845914 | 2.65 |
ENST00000373713.2
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr11_-_12030681 | 2.64 |
ENST00000529338.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chrX_+_152086373 | 2.64 |
ENST00000318529.8
|
ZNF185
|
zinc finger protein 185 (LIM domain) |
| chr1_+_3388181 | 2.61 |
ENST00000418137.1
ENST00000413250.2 |
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr22_+_31644388 | 2.60 |
ENST00000333611.4
ENST00000340552.4 |
LIMK2
|
LIM domain kinase 2 |
| chr5_+_9546376 | 2.59 |
ENST00000509788.1
|
SNHG18
|
small nucleolar RNA host gene 18 (non-protein coding) |
| chr9_+_124413873 | 2.58 |
ENST00000408936.3
|
DAB2IP
|
DAB2 interacting protein |
| chr19_+_49713991 | 2.56 |
ENST00000597316.1
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr14_+_72052983 | 2.54 |
ENST00000358550.2
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr1_+_150521876 | 2.53 |
ENST00000369041.5
ENST00000271643.4 ENST00000538795.1 |
ADAMTSL4
AL356356.1
|
ADAMTS-like 4 Protein LOC100996516 |
| chr19_+_52873166 | 2.49 |
ENST00000424032.2
ENST00000600321.1 ENST00000344085.5 ENST00000597976.1 ENST00000422689.2 |
ZNF880
|
zinc finger protein 880 |
| chr11_+_46403303 | 2.48 |
ENST00000407067.1
ENST00000395565.1 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr17_-_34122596 | 2.48 |
ENST00000250144.8
|
MMP28
|
matrix metallopeptidase 28 |
| chrX_-_7895479 | 2.47 |
ENST00000381042.4
|
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr8_+_94929168 | 2.45 |
ENST00000518107.1
ENST00000396200.3 |
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr7_-_994302 | 2.44 |
ENST00000265846.5
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr10_+_88718397 | 2.44 |
ENST00000372017.3
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1) |
| chr3_-_128840604 | 2.44 |
ENST00000476465.1
ENST00000315150.5 ENST00000393304.1 ENST00000393308.1 ENST00000393307.1 ENST00000393305.1 |
RAB43
|
RAB43, member RAS oncogene family |
| chr11_+_5712234 | 2.43 |
ENST00000414641.1
|
TRIM22
|
tripartite motif containing 22 |
| chr1_-_22215192 | 2.42 |
ENST00000374673.3
|
HSPG2
|
heparan sulfate proteoglycan 2 |
| chr9_-_122131696 | 2.41 |
ENST00000373964.2
ENST00000265922.3 |
BRINP1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr8_+_94929077 | 2.41 |
ENST00000297598.4
ENST00000520614.1 |
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr11_-_12030130 | 2.40 |
ENST00000450094.2
ENST00000534511.1 |
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr19_+_38810447 | 2.40 |
ENST00000263372.3
|
KCNK6
|
potassium channel, subfamily K, member 6 |
| chr1_-_95285764 | 2.37 |
ENST00000414374.1
ENST00000421997.1 ENST00000418366.2 ENST00000452922.1 |
LINC01057
|
long intergenic non-protein coding RNA 1057 |
| chr9_-_101017862 | 2.37 |
ENST00000375064.1
ENST00000342112.5 |
TBC1D2
|
TBC1 domain family, member 2 |
| chr1_+_150522222 | 2.35 |
ENST00000369039.5
|
ADAMTSL4
|
ADAMTS-like 4 |
| chr14_-_24729251 | 2.32 |
ENST00000559136.1
|
TGM1
|
transglutaminase 1 |
| chr3_-_176914998 | 2.32 |
ENST00000431421.1
ENST00000422066.1 ENST00000413084.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr19_+_48281803 | 2.31 |
ENST00000601048.1
|
SEPW1
|
selenoprotein W, 1 |
| chr21_-_47604318 | 2.29 |
ENST00000291672.5
ENST00000330205.6 |
SPATC1L
|
spermatogenesis and centriole associated 1-like |
| chr21_-_45079341 | 2.27 |
ENST00000443485.1
ENST00000291560.2 |
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chr9_-_101017900 | 2.26 |
ENST00000375066.5
|
TBC1D2
|
TBC1 domain family, member 2 |
| chr19_-_51522955 | 2.25 |
ENST00000358789.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr11_-_12030629 | 2.24 |
ENST00000396505.2
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr11_-_568369 | 2.23 |
ENST00000534540.1
ENST00000528245.1 ENST00000500447.1 ENST00000533920.1 |
MIR210HG
|
MIR210 host gene (non-protein coding) |
| chr22_+_25465786 | 2.23 |
ENST00000401395.1
|
KIAA1671
|
KIAA1671 |
| chr15_-_72490114 | 2.22 |
ENST00000309731.7
|
GRAMD2
|
GRAM domain containing 2 |
| chr8_+_94929110 | 2.21 |
ENST00000520728.1
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr3_+_38035610 | 2.18 |
ENST00000465644.1
|
VILL
|
villin-like |
| chr16_+_4526341 | 2.17 |
ENST00000458134.3
ENST00000219700.6 ENST00000575120.1 ENST00000572812.1 ENST00000574466.1 ENST00000576827.1 ENST00000570445.1 |
HMOX2
|
heme oxygenase (decycling) 2 |
| chr1_+_156123318 | 2.13 |
ENST00000368285.3
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr3_-_176915036 | 2.13 |
ENST00000427349.1
ENST00000352800.5 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr20_-_50808236 | 2.11 |
ENST00000361387.2
|
ZFP64
|
ZFP64 zinc finger protein |
| chr5_+_150157444 | 2.10 |
ENST00000526627.1
|
SMIM3
|
small integral membrane protein 3 |
| chr11_+_32851487 | 2.10 |
ENST00000257836.3
|
PRRG4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr6_+_43739697 | 2.10 |
ENST00000230480.6
|
VEGFA
|
vascular endothelial growth factor A |
| chr21_-_33985127 | 2.09 |
ENST00000290155.3
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr6_-_134373732 | 2.08 |
ENST00000275230.5
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr1_+_26605618 | 2.08 |
ENST00000270792.5
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr19_+_21688366 | 2.07 |
ENST00000358491.4
ENST00000597078.1 |
ZNF429
|
zinc finger protein 429 |
| chr16_+_3115611 | 2.07 |
ENST00000530890.1
ENST00000444393.3 ENST00000533097.2 ENST00000008180.9 ENST00000396890.2 ENST00000525228.1 ENST00000548652.1 ENST00000525377.2 ENST00000530538.2 ENST00000549213.1 ENST00000552936.1 ENST00000548476.1 ENST00000552664.1 ENST00000552356.1 ENST00000551513.1 ENST00000382213.3 ENST00000548246.1 |
IL32
|
interleukin 32 |
| chr12_-_8814669 | 2.06 |
ENST00000535411.1
ENST00000540087.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr2_-_85636928 | 2.06 |
ENST00000449030.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr3_-_16554403 | 2.06 |
ENST00000449415.1
ENST00000441460.1 |
RFTN1
|
raftlin, lipid raft linker 1 |
| chr19_-_2051223 | 2.04 |
ENST00000309340.7
ENST00000589534.1 ENST00000250896.3 ENST00000589509.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr1_+_203595903 | 2.04 |
ENST00000367218.3
ENST00000367219.3 ENST00000391954.2 |
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr1_+_36554470 | 2.01 |
ENST00000373178.4
|
ADPRHL2
|
ADP-ribosylhydrolase like 2 |
| chr11_-_118789613 | 1.99 |
ENST00000532899.1
|
BCL9L
|
B-cell CLL/lymphoma 9-like |
| chr21_-_33984888 | 1.98 |
ENST00000382549.4
ENST00000540881.1 |
C21orf59
|
chromosome 21 open reading frame 59 |
| chr8_-_119634141 | 1.98 |
ENST00000409003.4
ENST00000526328.1 ENST00000314727.4 ENST00000526765.1 |
SAMD12
|
sterile alpha motif domain containing 12 |
| chr12_-_13529642 | 1.98 |
ENST00000318426.2
|
C12orf36
|
chromosome 12 open reading frame 36 |
| chr17_+_36584662 | 1.97 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr12_-_120687948 | 1.97 |
ENST00000458477.2
|
PXN
|
paxillin |
| chr17_+_8339340 | 1.97 |
ENST00000580012.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr20_-_6104191 | 1.93 |
ENST00000217289.4
|
FERMT1
|
fermitin family member 1 |
| chr12_-_48276710 | 1.93 |
ENST00000550314.1
|
VDR
|
vitamin D (1,25- dihydroxyvitamin D3) receptor |
| chr11_-_65640198 | 1.93 |
ENST00000528176.1
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2 |
| chr12_-_54813229 | 1.92 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr1_-_20834586 | 1.92 |
ENST00000264198.3
|
MUL1
|
mitochondrial E3 ubiquitin protein ligase 1 |
| chr17_-_4463856 | 1.91 |
ENST00000574584.1
ENST00000381550.3 ENST00000301395.3 |
GGT6
|
gamma-glutamyltransferase 6 |
| chr19_-_20748541 | 1.91 |
ENST00000427401.4
ENST00000594419.1 |
ZNF737
|
zinc finger protein 737 |
| chr16_+_27325202 | 1.90 |
ENST00000395762.2
ENST00000562142.1 ENST00000561742.1 ENST00000543915.2 ENST00000449195.1 ENST00000380922.3 ENST00000563002.1 |
IL4R
|
interleukin 4 receptor |
| chr11_-_13484713 | 1.90 |
ENST00000526841.1
ENST00000529708.1 ENST00000278174.5 ENST00000528120.1 |
BTBD10
|
BTB (POZ) domain containing 10 |
| chr19_-_23578220 | 1.90 |
ENST00000595533.1
ENST00000397082.2 ENST00000599743.1 ENST00000300619.7 |
ZNF91
|
zinc finger protein 91 |
| chr9_+_112542591 | 1.90 |
ENST00000483909.1
ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2
PALM2-AKAP2
AKAP2
|
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr5_-_42825983 | 1.89 |
ENST00000506577.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr9_-_139891165 | 1.89 |
ENST00000494426.1
|
CLIC3
|
chloride intracellular channel 3 |
| chr9_+_135937365 | 1.89 |
ENST00000372080.4
ENST00000351304.7 |
CEL
|
carboxyl ester lipase |
| chr14_+_94577074 | 1.88 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr15_-_101084547 | 1.87 |
ENST00000394113.1
|
CERS3
|
ceramide synthase 3 |
| chr2_+_131769256 | 1.86 |
ENST00000355771.3
|
ARHGEF4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr19_-_2041159 | 1.86 |
ENST00000589441.1
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr10_-_97050777 | 1.86 |
ENST00000329399.6
|
PDLIM1
|
PDZ and LIM domain 1 |
| chr13_-_20806440 | 1.85 |
ENST00000400066.3
ENST00000400065.3 ENST00000356192.6 |
GJB6
|
gap junction protein, beta 6, 30kDa |
| chr6_+_27356745 | 1.83 |
ENST00000461521.1
|
ZNF391
|
zinc finger protein 391 |
| chr11_+_111412053 | 1.83 |
ENST00000530962.1
|
LAYN
|
layilin |
| chr11_-_111784005 | 1.82 |
ENST00000527899.1
|
CRYAB
|
crystallin, alpha B |
| chr13_+_115047097 | 1.82 |
ENST00000351487.5
|
UPF3A
|
UPF3 regulator of nonsense transcripts homolog A (yeast) |
| chr9_-_34458531 | 1.81 |
ENST00000379089.1
ENST00000379087.1 ENST00000379084.1 ENST00000379081.1 ENST00000379080.1 ENST00000422409.1 ENST00000379078.1 ENST00000445726.1 ENST00000297620.4 |
FAM219A
|
family with sequence similarity 219, member A |
| chr1_+_220701740 | 1.80 |
ENST00000366917.4
|
MARK1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr14_+_29236269 | 1.79 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr11_+_18230727 | 1.77 |
ENST00000527059.1
|
RP11-113D6.10
|
Putative mitochondrial carrier protein LOC494141 |
| chr11_-_111783595 | 1.77 |
ENST00000528628.1
|
CRYAB
|
crystallin, alpha B |
| chr6_-_33548006 | 1.75 |
ENST00000374467.3
|
BAK1
|
BCL2-antagonist/killer 1 |
| chr4_-_177713788 | 1.75 |
ENST00000280193.2
|
VEGFC
|
vascular endothelial growth factor C |
| chr2_-_264772 | 1.75 |
ENST00000403658.1
ENST00000402632.1 ENST00000415368.1 ENST00000454318.1 |
SH3YL1
|
SH3 and SYLF domain containing 1 |
| chr12_-_54778471 | 1.75 |
ENST00000550120.1
ENST00000394313.2 ENST00000547210.1 |
ZNF385A
|
zinc finger protein 385A |
| chr8_+_96146168 | 1.75 |
ENST00000519516.1
|
PLEKHF2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr19_+_2096868 | 1.73 |
ENST00000395296.1
ENST00000395301.3 |
IZUMO4
|
IZUMO family member 4 |
| chr10_+_11060004 | 1.73 |
ENST00000542579.1
ENST00000399850.3 ENST00000417956.2 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr2_-_201936302 | 1.73 |
ENST00000453765.1
ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B
|
family with sequence similarity 126, member B |
| chr11_+_46403194 | 1.72 |
ENST00000395569.4
ENST00000395566.4 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chrX_-_52683950 | 1.71 |
ENST00000298181.5
|
SSX7
|
synovial sarcoma, X breakpoint 7 |
| chr1_-_152386732 | 1.71 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr3_+_186648307 | 1.71 |
ENST00000457772.2
ENST00000455441.1 ENST00000427315.1 |
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr12_-_57030115 | 1.70 |
ENST00000379441.3
ENST00000179765.5 ENST00000551812.1 |
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr1_+_40420802 | 1.70 |
ENST00000372811.5
ENST00000420632.2 ENST00000434861.1 ENST00000372809.5 |
MFSD2A
|
major facilitator superfamily domain containing 2A |
| chr6_-_167040693 | 1.69 |
ENST00000366863.2
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GLI3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 12.1 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 2.9 | 11.6 | GO:1902910 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 2.5 | 15.1 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 1.8 | 3.6 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 1.7 | 8.4 | GO:2000470 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 1.6 | 4.8 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 1.4 | 4.1 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 1.2 | 8.5 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 1.1 | 3.4 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 1.1 | 3.4 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 1.0 | 5.0 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 1.0 | 6.0 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 1.0 | 5.7 | GO:0042335 | cuticle development(GO:0042335) |
| 0.9 | 6.6 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.9 | 7.3 | GO:1902613 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.9 | 3.5 | GO:1903381 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 0.9 | 5.2 | GO:0030421 | defecation(GO:0030421) |
| 0.9 | 4.3 | GO:0032796 | uropod organization(GO:0032796) |
| 0.8 | 2.5 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.8 | 2.4 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.8 | 4.0 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.8 | 3.8 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.7 | 2.1 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.7 | 2.0 | GO:1900081 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.6 | 24.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.6 | 1.9 | GO:1903521 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.6 | 2.6 | GO:1990736 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.6 | 1.9 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.6 | 2.9 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.6 | 2.8 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.6 | 5.7 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.5 | 2.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.5 | 2.7 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.5 | 0.5 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.5 | 2.6 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.5 | 1.5 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.5 | 4.5 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.5 | 1.5 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.5 | 2.9 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.5 | 2.9 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.5 | 1.9 | GO:0070895 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.5 | 1.9 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.4 | 0.9 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.4 | 1.7 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.4 | 2.5 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.4 | 3.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.4 | 2.7 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.4 | 2.3 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.4 | 2.7 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.4 | 5.7 | GO:0060613 | fat pad development(GO:0060613) |
| 0.4 | 1.9 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.4 | 2.3 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.4 | 1.9 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.4 | 3.3 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.4 | 1.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.4 | 0.7 | GO:0006913 | nucleocytoplasmic transport(GO:0006913) |
| 0.4 | 1.5 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.4 | 4.0 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.4 | 6.8 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.3 | 1.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 2.8 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.3 | 6.2 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.3 | 1.4 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.3 | 2.7 | GO:0030578 | PML body organization(GO:0030578) |
| 0.3 | 4.4 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.3 | 0.7 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.3 | 1.0 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.3 | 2.3 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.3 | 3.0 | GO:0003383 | apical constriction(GO:0003383) |
| 0.3 | 2.9 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.3 | 0.9 | GO:0046707 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.3 | 1.3 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.3 | 0.9 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.3 | 4.6 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.3 | 1.5 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.3 | 1.2 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.3 | 1.8 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.3 | 2.3 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.3 | 8.2 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.3 | 0.8 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.3 | 3.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.3 | 1.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.3 | 7.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.3 | 2.3 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.3 | 1.3 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.3 | 1.5 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 1.5 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.2 | 3.0 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 1.5 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.2 | 2.9 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.2 | 1.0 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.2 | 1.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.2 | 4.8 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 0.7 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.2 | 0.9 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 1.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 0.7 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 1.3 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.2 | 0.7 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.2 | 0.9 | GO:0030262 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.2 | 0.9 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.2 | 0.6 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.2 | 1.5 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.2 | 5.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 4.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 2.5 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 6.7 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.2 | 1.9 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.2 | 0.6 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.2 | 1.2 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.2 | 0.6 | GO:0030451 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 0.2 | 1.2 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 | 2.3 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.2 | 0.6 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.2 | 1.3 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.2 | 1.5 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.2 | 0.9 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.2 | 5.5 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.2 | 0.7 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.2 | 4.5 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.2 | 1.6 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.2 | 2.0 | GO:2000467 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.2 | 0.5 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.2 | 1.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.2 | 1.0 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.2 | 0.8 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.2 | 3.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 0.5 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.2 | 4.0 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.2 | 1.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.2 | 3.9 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 | 0.9 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.2 | 1.9 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.2 | 0.9 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.2 | 1.4 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 2.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 0.3 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 1.6 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.4 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.1 | 0.4 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.1 | 1.0 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 2.9 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 0.7 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.4 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 1.6 | GO:1904847 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.1 | 1.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 2.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 1.4 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 1.8 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 1.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 2.4 | GO:0007614 | short-term memory(GO:0007614) |
| 0.1 | 1.5 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.5 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 4.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.5 | GO:0002678 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) positive regulation of chronic inflammatory response(GO:0002678) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.1 | 0.8 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.1 | 5.3 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 0.4 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.1 | 0.6 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 1.1 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 4.4 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 1.0 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.1 | 3.0 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.1 | 5.2 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 1.3 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 | 1.3 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.1 | 0.3 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 2.3 | GO:0070831 | basement membrane assembly(GO:0070831) regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 1.6 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 1.5 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.5 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 1.3 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 0.2 | GO:0072107 | cranial ganglion development(GO:0061550) trigeminal ganglion development(GO:0061551) regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.1 | 5.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 1.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 1.9 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 0.3 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.9 | GO:0006868 | glutamine transport(GO:0006868) L-cystine transport(GO:0015811) |
| 0.1 | 2.9 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 1.1 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.1 | 0.4 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.1 | 0.4 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 3.6 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.6 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 1.1 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) |
| 0.1 | 0.3 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.1 | 2.1 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.1 | 0.8 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 1.7 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.1 | 0.6 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.7 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.1 | 0.7 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.1 | 3.8 | GO:0043090 | amino acid import(GO:0043090) |
| 0.1 | 2.0 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.1 | 0.4 | GO:0030011 | maintenance of cell polarity(GO:0030011) maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.8 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.1 | 1.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 2.1 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 0.8 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 2.2 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 1.0 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.6 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 1.9 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 1.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.3 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.1 | 1.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.2 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.1 | 1.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.3 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.1 | 0.4 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.1 | 0.8 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 1.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.4 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.1 | 1.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 1.0 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 1.9 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 0.9 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.1 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.1 | 0.9 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.7 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 0.6 | GO:0046349 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.1 | 0.4 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 0.2 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.1 | 1.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.9 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 0.6 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 0.5 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 0.7 | GO:0051583 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.1 | 1.4 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 1.0 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.4 | GO:0006311 | meiotic gene conversion(GO:0006311) |
| 0.1 | 19.6 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.1 | 0.5 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 0.4 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 1.2 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.1 | 3.7 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.8 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.1 | 0.5 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 2.5 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.1 | 0.4 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 2.8 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 0.3 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 1.9 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.5 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 0.7 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.5 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 1.8 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 0.1 | GO:0014009 | glial cell proliferation(GO:0014009) |
| 0.1 | 0.4 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.1 | 0.3 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.1 | 7.2 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 1.2 | GO:0030208 | dermatan sulfate metabolic process(GO:0030205) dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.7 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 0.4 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.1 | 1.5 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 0.1 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 3.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 2.6 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.5 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.7 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 1.4 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 1.1 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 1.8 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 0.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.7 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 0.1 | 0.4 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 1.7 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 0.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 2.0 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 0.5 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.2 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 | 0.5 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.1 | 1.0 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.1 | 0.6 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 1.7 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 3.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.9 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 0.8 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 7.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 1.8 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 1.1 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 1.0 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.1 | 0.4 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.5 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 2.5 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.2 | GO:0090410 | malonate catabolic process(GO:0090410) |
| 0.0 | 0.6 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 1.7 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 3.1 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.4 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.4 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.8 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.5 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.0 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.0 | 1.1 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.6 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.2 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 2.8 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.1 | GO:0050823 | peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.0 | 0.9 | GO:0050890 | cognition(GO:0050890) |
| 0.0 | 0.5 | GO:0048839 | inner ear development(GO:0048839) |
| 0.0 | 0.7 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 2.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.9 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 7.5 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.6 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 1.7 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.7 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 1.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 2.7 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.3 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.5 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.9 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 | 0.4 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 2.8 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.1 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.0 | 0.1 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 1.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.2 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 3.4 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.3 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 2.7 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.5 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.8 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.4 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 0.2 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 1.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.5 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.4 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.2 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.2 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.5 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 2.3 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 1.6 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.3 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.2 | GO:0019556 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.6 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.0 | 0.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.6 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 1.2 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.3 | GO:0015871 | choline transport(GO:0015871) negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 1.9 | GO:0006901 | vesicle coating(GO:0006901) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.3 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.2 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.2 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 1.8 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 1.0 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.0 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.2 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.9 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0051918 | regulation of fibrinolysis(GO:0051917) negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.3 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.6 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.0 | 0.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.9 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 0.1 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.4 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) |
| 0.0 | 0.5 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 1.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.2 | GO:0060180 | female mating behavior(GO:0060180) |
| 0.0 | 0.4 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.8 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) |
| 0.0 | 0.7 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.3 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.2 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.9 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.0 | 0.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.7 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.3 | GO:0046320 | regulation of fatty acid oxidation(GO:0046320) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.7 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 1.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.1 | GO:1901203 | positive regulation of extracellular matrix assembly(GO:1901203) |
| 0.0 | 0.1 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.0 | 0.4 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 15.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 2.1 | 8.4 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 1.9 | 7.7 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 1.9 | 1.9 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 1.2 | 13.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 1.1 | 3.4 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.9 | 2.6 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.6 | 4.5 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.6 | 3.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.5 | 3.8 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.5 | 2.8 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.4 | 1.1 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.3 | 6.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.3 | 2.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.3 | 8.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.3 | 4.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.3 | 1.8 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.3 | 4.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 0.9 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.3 | 1.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.3 | 22.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 1.5 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.2 | 9.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 4.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 0.7 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 1.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.2 | 0.7 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.2 | 3.7 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 0.9 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.2 | 1.6 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 2.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 1.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 1.6 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 3.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 1.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 0.6 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.2 | 1.1 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.2 | 0.9 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.2 | 0.9 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.2 | 1.0 | GO:0071546 | pi-body(GO:0071546) |
| 0.2 | 0.9 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.2 | 0.5 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.2 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 3.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 2.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 0.8 | GO:0071547 | piP-body(GO:0071547) |
| 0.2 | 0.5 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.2 | 11.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 2.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.2 | 4.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 1.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.2 | 0.6 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 0.9 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 2.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 4.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 2.4 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 1.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.8 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.1 | 7.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.5 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 1.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 2.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 1.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.6 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 2.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 2.8 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.3 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 10.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.9 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 1.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 2.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 2.1 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.4 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 4.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 0.8 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.1 | 1.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.4 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.1 | 4.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.9 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 9.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 2.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 0.3 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 1.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.3 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.1 | 3.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 10.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.3 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.1 | 2.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 1.7 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 2.3 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 7.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 9.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.7 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 5.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 2.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.6 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 0.6 | GO:0005694 | chromosome(GO:0005694) |
| 0.1 | 2.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 2.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.5 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 8.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 4.5 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.7 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 4.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 2.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.4 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 1.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 1.6 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 1.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.5 | GO:0030681 | multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 2.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.5 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 5.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.9 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 1.9 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 1.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 3.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.3 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 1.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 2.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 3.2 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.2 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 3.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.4 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.6 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 3.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 13.8 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 3.3 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 0.7 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.9 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 1.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0071556 | integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 1.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.0 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 1.4 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 1.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.4 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 1.0 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.7 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 1.8 | 9.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 1.7 | 8.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 1.2 | 7.3 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 1.1 | 5.3 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 1.0 | 7.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 1.0 | 2.9 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.9 | 2.7 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.7 | 3.0 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.7 | 2.9 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) |
| 0.7 | 2.0 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.6 | 1.9 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.6 | 1.9 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.6 | 3.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.6 | 1.7 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.5 | 2.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.5 | 11.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.5 | 1.6 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.5 | 4.5 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.5 | 1.5 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.4 | 1.3 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.4 | 5.8 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.4 | 5.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.4 | 1.1 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.4 | 1.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.3 | 2.8 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.3 | 4.8 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.3 | 1.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.3 | 8.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.3 | 0.9 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.3 | 0.9 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.3 | 2.7 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.3 | 2.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.3 | 2.9 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.3 | 0.9 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.3 | 3.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.3 | 1.1 | GO:0031716 | calcitonin receptor activity(GO:0004948) calcitonin receptor binding(GO:0031716) |
| 0.3 | 1.4 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.3 | 1.4 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.3 | 1.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.3 | 1.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.3 | 1.5 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.3 | 3.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.3 | 1.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 1.5 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.2 | 4.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 1.0 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.2 | 1.9 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.2 | 0.9 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.2 | 0.9 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.2 | 2.4 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.2 | 2.4 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.2 | 2.8 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.2 | 5.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 6.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 0.6 | GO:0004877 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.2 | 3.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 4.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 0.9 | GO:0019144 | ADP-sugar diphosphatase activity(GO:0019144) |
| 0.2 | 1.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 3.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 1.9 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 0.9 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 3.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 0.5 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.2 | 3.9 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.2 | 0.7 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.2 | 3.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 0.6 | GO:0005334 | norepinephrine:sodium symporter activity(GO:0005334) |
| 0.2 | 3.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 0.5 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.4 | GO:0019961 | interferon binding(GO:0019961) |
| 0.1 | 0.7 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 1.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 2.5 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.4 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.1 | 2.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 2.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 1.6 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 2.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 2.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.6 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.1 | 2.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 3.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 1.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 3.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 1.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.9 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 0.9 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 2.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 3.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 1.6 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 1.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 1.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.5 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 15.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.5 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 3.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 1.1 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.0 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 1.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 11.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.9 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.3 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.1 | 1.1 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 2.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 3.1 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 0.4 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.1 | 1.9 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 1.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.9 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 2.0 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 2.7 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.6 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 21.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 3.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.7 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) maltose alpha-glucosidase activity(GO:0032450) |
| 0.1 | 0.5 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 2.4 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.1 | 0.2 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 0.5 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 10.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 2.0 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 3.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 1.5 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 3.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.6 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 16.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.6 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) |
| 0.1 | 0.4 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 6.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 3.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.3 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.1 | 1.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 2.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.3 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.6 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.8 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 3.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 0.6 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.2 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.1 | 0.9 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 1.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.7 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 1.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 2.7 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 1.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.8 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 3.2 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 2.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 1.0 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.1 | 0.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.3 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.1 | 0.9 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 0.5 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.1 | 0.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 3.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 4.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.6 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 1.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.0 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.2 | GO:0090409 | malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 0.6 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.8 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.2 | GO:0047977 | linoleate 13S-lipoxygenase activity(GO:0016165) hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 0.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 6.6 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 1.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 1.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 1.5 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.4 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.2 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.6 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 1.0 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 3.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.7 | GO:0070008 | serine-type carboxypeptidase activity(GO:0004185) serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.3 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 2.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.4 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 1.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 1.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 22.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 2.7 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.0 | GO:0098847 | sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 2.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.8 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 0.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 5.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 3.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 1.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.7 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 1.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.5 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.9 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.7 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 1.0 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 3.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 3.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.0 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 1.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 23.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 4.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 1.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.2 | 22.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 12.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 11.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 13.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 3.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 1.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 9.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 5.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 1.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 8.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 5.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 8.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.9 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 1.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 4.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 18.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 0.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 1.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 3.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 2.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 3.3 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 6.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 4.0 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 8.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.9 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.8 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.5 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 7.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.9 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.5 | 0.5 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.4 | 24.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 6.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 3.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 8.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.3 | 1.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.3 | 11.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 25.0 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.2 | 7.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 0.9 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 3.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 1.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.2 | 1.5 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 4.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.9 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 2.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 3.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 5.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 7.5 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 2.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 2.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.3 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 0.8 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 0.7 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 1.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 5.1 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 1.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 0.9 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 0.8 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.1 | 1.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 2.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 3.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 2.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 4.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 2.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 2.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 2.1 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 1.7 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 4.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.9 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 2.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.2 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 1.5 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.0 | 2.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 5.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 2.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 4.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.0 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.6 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 2.2 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 11.3 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 1.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.1 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |