Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for GLIS3
Z-value: 0.65
Transcription factors associated with GLIS3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLIS3
|
ENSG00000107249.17 | GLIS family zinc finger 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLIS3 | hg19_v2_chr9_-_4299874_4299916 | 0.96 | 1.4e-11 | Click! |
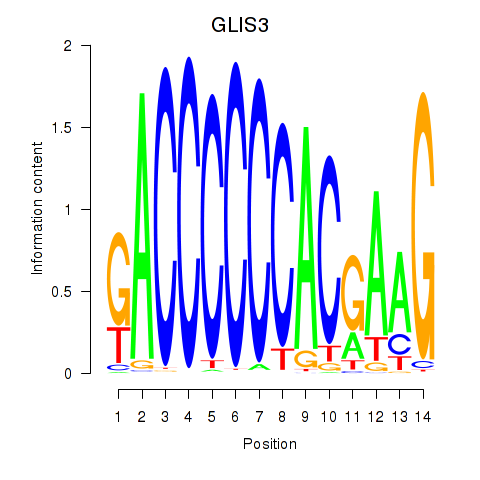
Activity profile of GLIS3 motif
Sorted Z-values of GLIS3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_247095236 | 1.93 |
ENST00000478568.1
|
AHCTF1
|
AT hook containing transcription factor 1 |
| chr19_+_48828582 | 1.54 |
ENST00000270221.6
ENST00000596315.1 |
EMP3
|
epithelial membrane protein 3 |
| chr21_+_38071430 | 1.51 |
ENST00000290399.6
|
SIM2
|
single-minded family bHLH transcription factor 2 |
| chr7_+_150758304 | 1.14 |
ENST00000482950.1
ENST00000463414.1 ENST00000310317.5 |
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr17_-_33446820 | 0.99 |
ENST00000592577.1
ENST00000590016.1 ENST00000345365.6 ENST00000360276.3 ENST00000357906.3 |
RAD51D
|
RAD51 paralog D |
| chr7_+_150758642 | 0.98 |
ENST00000488420.1
|
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr19_-_10613862 | 0.94 |
ENST00000592055.1
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr1_-_45988542 | 0.89 |
ENST00000424390.1
|
PRDX1
|
peroxiredoxin 1 |
| chr12_+_109577202 | 0.87 |
ENST00000377848.3
ENST00000377854.5 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr19_-_10613361 | 0.86 |
ENST00000591039.1
ENST00000591419.1 |
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr6_+_44191290 | 0.83 |
ENST00000371755.3
ENST00000371740.5 ENST00000371731.1 ENST00000393841.1 |
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr11_+_68451943 | 0.81 |
ENST00000265643.3
|
GAL
|
galanin/GMAP prepropeptide |
| chr7_+_65958693 | 0.78 |
ENST00000445681.1
ENST00000452565.1 |
GS1-124K5.4
|
GS1-124K5.4 |
| chr20_+_61867235 | 0.78 |
ENST00000342412.6
ENST00000217169.3 |
BIRC7
|
baculoviral IAP repeat containing 7 |
| chr17_+_80818231 | 0.78 |
ENST00000576996.1
|
TBCD
|
tubulin folding cofactor D |
| chr6_+_24495067 | 0.76 |
ENST00000357578.3
ENST00000546278.1 ENST00000491546.1 |
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr19_-_48673552 | 0.74 |
ENST00000536218.1
ENST00000596549.1 |
LIG1
|
ligase I, DNA, ATP-dependent |
| chr9_+_131903916 | 0.72 |
ENST00000419582.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr9_+_131904295 | 0.71 |
ENST00000434095.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr3_-_160167540 | 0.69 |
ENST00000496222.1
ENST00000471396.1 ENST00000471155.1 ENST00000309784.4 |
TRIM59
|
tripartite motif containing 59 |
| chr7_-_50518022 | 0.69 |
ENST00000356889.4
ENST00000420829.1 ENST00000448788.1 ENST00000395556.2 ENST00000422854.1 ENST00000435566.1 ENST00000433017.1 |
FIGNL1
|
fidgetin-like 1 |
| chr6_+_44191507 | 0.68 |
ENST00000371724.1
ENST00000371713.1 |
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr9_+_131904233 | 0.65 |
ENST00000432651.1
ENST00000435132.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr5_+_41925325 | 0.63 |
ENST00000296812.2
ENST00000281623.3 ENST00000509134.1 |
FBXO4
|
F-box protein 4 |
| chr3_+_127348005 | 0.60 |
ENST00000342480.6
|
PODXL2
|
podocalyxin-like 2 |
| chr14_+_77228532 | 0.60 |
ENST00000167106.4
ENST00000554237.1 |
VASH1
|
vasohibin 1 |
| chr6_+_24495185 | 0.57 |
ENST00000348925.2
|
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chrX_+_118370288 | 0.57 |
ENST00000535419.1
|
PGRMC1
|
progesterone receptor membrane component 1 |
| chr19_+_40697514 | 0.56 |
ENST00000253055.3
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr11_+_393428 | 0.56 |
ENST00000533249.1
ENST00000527442.1 |
PKP3
|
plakophilin 3 |
| chr12_+_132312931 | 0.55 |
ENST00000360564.1
ENST00000545671.1 ENST00000545790.1 |
MMP17
|
matrix metallopeptidase 17 (membrane-inserted) |
| chr10_+_71389983 | 0.51 |
ENST00000373279.4
|
C10orf35
|
chromosome 10 open reading frame 35 |
| chr1_-_45987526 | 0.50 |
ENST00000372079.1
ENST00000262746.1 ENST00000447184.1 ENST00000319248.8 |
PRDX1
|
peroxiredoxin 1 |
| chr15_+_91478493 | 0.48 |
ENST00000418476.2
|
UNC45A
|
unc-45 homolog A (C. elegans) |
| chr11_+_1463728 | 0.46 |
ENST00000544817.1
|
BRSK2
|
BR serine/threonine kinase 2 |
| chr22_-_31742218 | 0.46 |
ENST00000266269.5
ENST00000405309.3 ENST00000351933.4 |
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr9_-_130742792 | 0.45 |
ENST00000373095.1
|
FAM102A
|
family with sequence similarity 102, member A |
| chr5_+_172386517 | 0.44 |
ENST00000519522.1
|
RPL26L1
|
ribosomal protein L26-like 1 |
| chr22_+_50628999 | 0.40 |
ENST00000395827.1
|
TRABD
|
TraB domain containing |
| chr16_+_30006615 | 0.38 |
ENST00000563197.1
|
INO80E
|
INO80 complex subunit E |
| chrX_+_118370211 | 0.38 |
ENST00000217971.7
|
PGRMC1
|
progesterone receptor membrane component 1 |
| chr1_-_92371839 | 0.37 |
ENST00000370399.2
|
TGFBR3
|
transforming growth factor, beta receptor III |
| chr11_-_68671264 | 0.35 |
ENST00000362034.2
|
MRPL21
|
mitochondrial ribosomal protein L21 |
| chr3_-_160167301 | 0.32 |
ENST00000494486.1
|
TRIM59
|
tripartite motif containing 59 |
| chr6_+_33172407 | 0.31 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr3_-_160167508 | 0.31 |
ENST00000479460.1
|
TRIM59
|
tripartite motif containing 59 |
| chr8_-_74791051 | 0.30 |
ENST00000453587.2
ENST00000602969.1 ENST00000602593.1 ENST00000419880.3 ENST00000517608.1 |
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr2_+_219824357 | 0.29 |
ENST00000302625.4
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39) |
| chr11_-_118789613 | 0.28 |
ENST00000532899.1
|
BCL9L
|
B-cell CLL/lymphoma 9-like |
| chr8_-_12051576 | 0.27 |
ENST00000524571.2
ENST00000533852.2 ENST00000533513.1 ENST00000448228.2 ENST00000534520.1 ENST00000321602.8 |
FAM86B1
|
family with sequence similarity 86, member B1 |
| chr12_+_48152774 | 0.26 |
ENST00000549243.1
|
SLC48A1
|
solute carrier family 48 (heme transporter), member 1 |
| chr4_-_84255935 | 0.25 |
ENST00000513463.1
|
HPSE
|
heparanase |
| chr8_-_29120580 | 0.23 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr20_+_18447771 | 0.22 |
ENST00000377603.4
|
POLR3F
|
polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa |
| chr5_+_172386419 | 0.21 |
ENST00000265100.2
ENST00000519239.1 |
RPL26L1
|
ribosomal protein L26-like 1 |
| chr11_+_68671310 | 0.21 |
ENST00000255078.3
ENST00000539224.1 |
IGHMBP2
|
immunoglobulin mu binding protein 2 |
| chr17_-_56605341 | 0.20 |
ENST00000583114.1
|
SEPT4
|
septin 4 |
| chr12_+_132568814 | 0.20 |
ENST00000454179.2
ENST00000389560.2 ENST00000443539.2 ENST00000392352.1 |
EP400NL
|
EP400 N-terminal like |
| chr9_-_98279241 | 0.20 |
ENST00000437951.1
ENST00000375274.2 ENST00000430669.2 ENST00000468211.2 |
PTCH1
|
patched 1 |
| chr8_-_29120604 | 0.20 |
ENST00000521515.1
|
KIF13B
|
kinesin family member 13B |
| chr3_-_25824925 | 0.19 |
ENST00000396649.3
ENST00000428257.1 ENST00000280700.5 |
NGLY1
|
N-glycanase 1 |
| chr17_-_77925806 | 0.15 |
ENST00000574241.2
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr19_-_56092187 | 0.13 |
ENST00000325421.4
ENST00000592239.1 |
ZNF579
|
zinc finger protein 579 |
| chr19_-_49828438 | 0.12 |
ENST00000454748.3
ENST00000598828.1 ENST00000335875.4 |
SLC6A16
|
solute carrier family 6, member 16 |
| chr20_-_44718538 | 0.12 |
ENST00000290231.6
ENST00000372291.3 |
NCOA5
|
nuclear receptor coactivator 5 |
| chr19_+_46850251 | 0.12 |
ENST00000012443.4
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr17_-_5015129 | 0.12 |
ENST00000575898.1
ENST00000416429.2 |
ZNF232
|
zinc finger protein 232 |
| chr1_+_110527308 | 0.12 |
ENST00000369799.5
|
AHCYL1
|
adenosylhomocysteinase-like 1 |
| chr3_+_173116225 | 0.11 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr7_+_23636992 | 0.10 |
ENST00000307471.3
ENST00000409765.1 |
CCDC126
|
coiled-coil domain containing 126 |
| chr5_-_137090028 | 0.08 |
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr22_-_50964849 | 0.08 |
ENST00000543927.1
ENST00000423348.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr8_+_11627148 | 0.08 |
ENST00000436750.3
|
NEIL2
|
nei endonuclease VIII-like 2 (E. coli) |
| chr11_+_5646213 | 0.07 |
ENST00000429814.2
|
TRIM34
|
tripartite motif containing 34 |
| chr19_+_1495362 | 0.07 |
ENST00000395479.4
|
REEP6
|
receptor accessory protein 6 |
| chr12_-_57400227 | 0.07 |
ENST00000300101.2
|
ZBTB39
|
zinc finger and BTB domain containing 39 |
| chr18_+_3451584 | 0.06 |
ENST00000551541.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr8_+_11627205 | 0.05 |
ENST00000455213.2
ENST00000403422.3 ENST00000528323.1 ENST00000284503.6 |
NEIL2
|
nei endonuclease VIII-like 2 (E. coli) |
| chr1_-_53019059 | 0.05 |
ENST00000484723.2
ENST00000524582.1 |
ZCCHC11
|
zinc finger, CCHC domain containing 11 |
| chr17_+_7792101 | 0.04 |
ENST00000358181.4
ENST00000330494.7 |
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr11_-_8680383 | 0.04 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chr18_+_3451646 | 0.04 |
ENST00000345133.5
ENST00000330513.5 ENST00000549546.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr11_+_57308979 | 0.03 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr19_+_46850320 | 0.03 |
ENST00000391919.1
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr3_-_25824872 | 0.03 |
ENST00000308710.5
|
NGLY1
|
N-glycanase 1 |
| chr17_+_5389605 | 0.03 |
ENST00000576988.1
ENST00000576570.1 ENST00000573759.1 |
MIS12
|
MIS12 kinetochore complex component |
| chr4_-_74904398 | 0.03 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chr22_+_18834324 | 0.02 |
ENST00000342005.4
|
AC008132.13
|
Uncharacterized protein |
| chr2_-_241737128 | 0.01 |
ENST00000404283.3
|
KIF1A
|
kinesin family member 1A |
| chr16_+_89686991 | 0.01 |
ENST00000393092.3
|
DPEP1
|
dipeptidase 1 (renal) |
| chr1_+_1950763 | 0.01 |
ENST00000378585.4
|
GABRD
|
gamma-aminobutyric acid (GABA) A receptor, delta |
| chr17_-_79917645 | 0.00 |
ENST00000477214.1
|
NOTUM
|
notum pectinacetylesterase homolog (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of GLIS3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0015862 | uridine transport(GO:0015862) |
| 0.3 | 0.8 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.3 | 1.3 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.2 | 0.6 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.9 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 1.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 1.0 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 0.3 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 1.9 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.4 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.8 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.6 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.6 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.6 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 1.5 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.6 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 1.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.2 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 1.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.2 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.8 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.4 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 2.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.5 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.0 | 0.6 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.7 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 1.5 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 1.9 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.4 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 1.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 1.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0097227 | sperm annulus(GO:0097227) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.6 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 1.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.8 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.4 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.1 | 0.3 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.1 | 0.3 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 2.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.2 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.1 | 0.2 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 1.0 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 1.5 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.2 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.3 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 2.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.9 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 2.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |