Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
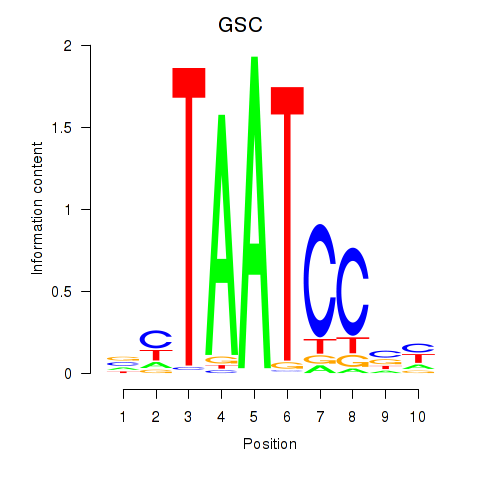
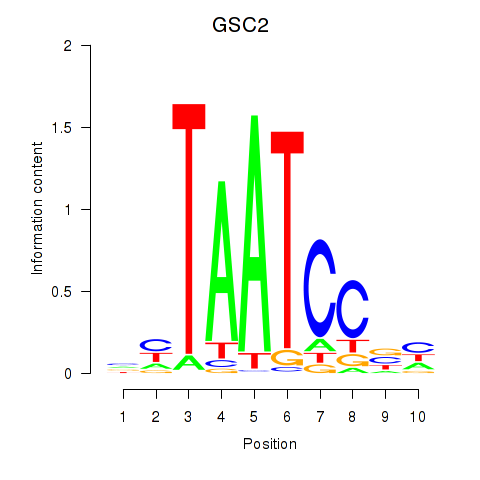
Results for GSC_GSC2
Z-value: 0.73
Transcription factors associated with GSC_GSC2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GSC
|
ENSG00000133937.3 | goosecoid homeobox |
|
GSC2
|
ENSG00000063515.2 | goosecoid homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GSC | hg19_v2_chr14_-_95236551_95236576 | 0.69 | 8.1e-04 | Click! |
| GSC2 | hg19_v2_chr22_-_19137796_19137796 | -0.23 | 3.2e-01 | Click! |
Activity profile of GSC_GSC2 motif
Sorted Z-values of GSC_GSC2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_61522844 | 3.09 |
ENST00000265460.5
|
MYRF
|
myelin regulatory factor |
| chr15_-_27184664 | 2.83 |
ENST00000541819.2
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr12_-_49523896 | 2.33 |
ENST00000549870.1
|
TUBA1B
|
tubulin, alpha 1b |
| chr16_+_81272287 | 1.88 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr2_-_175711133 | 1.87 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr17_-_19651668 | 1.52 |
ENST00000494157.2
ENST00000225740.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr17_-_19651654 | 1.48 |
ENST00000395555.3
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr5_+_76114758 | 1.46 |
ENST00000514165.1
ENST00000296677.4 |
F2RL1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr10_+_104178946 | 1.43 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr1_+_153651078 | 1.41 |
ENST00000368680.3
|
NPR1
|
natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A) |
| chr3_-_149388682 | 1.41 |
ENST00000475579.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr17_-_19651598 | 1.39 |
ENST00000570414.1
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr19_+_50183032 | 1.26 |
ENST00000527412.1
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr1_-_47655686 | 1.16 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chrX_-_10588595 | 1.12 |
ENST00000423614.1
ENST00000317552.4 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr3_+_111393501 | 1.10 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr13_-_31191642 | 1.09 |
ENST00000405805.1
|
HMGB1
|
high mobility group box 1 |
| chr15_+_63050785 | 1.06 |
ENST00000472902.1
|
TLN2
|
talin 2 |
| chr9_+_103204553 | 1.02 |
ENST00000502978.1
ENST00000334943.6 |
MSANTD3-TMEFF1
TMEFF1
|
MSANTD3-TMEFF1 readthrough transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chrX_+_153769446 | 1.02 |
ENST00000422680.1
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr12_+_7053172 | 0.96 |
ENST00000229281.5
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr12_+_78224667 | 0.95 |
ENST00000549464.1
|
NAV3
|
neuron navigator 3 |
| chr16_-_31076332 | 0.94 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr1_+_17248418 | 0.92 |
ENST00000375541.5
|
CROCC
|
ciliary rootlet coiled-coil, rootletin |
| chr17_-_4689649 | 0.89 |
ENST00000441199.2
ENST00000416307.2 |
VMO1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chrX_+_69509927 | 0.86 |
ENST00000374403.3
|
KIF4A
|
kinesin family member 4A |
| chr1_+_200863949 | 0.85 |
ENST00000413687.2
|
C1orf106
|
chromosome 1 open reading frame 106 |
| chr15_-_52483566 | 0.85 |
ENST00000261837.7
|
GNB5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr14_-_101034407 | 0.85 |
ENST00000443071.2
ENST00000557378.1 |
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr2_-_175499294 | 0.85 |
ENST00000392547.2
|
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chrX_+_69509870 | 0.84 |
ENST00000374388.3
|
KIF4A
|
kinesin family member 4A |
| chrX_-_10588459 | 0.83 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr9_-_131486367 | 0.82 |
ENST00000372663.4
ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12
|
zinc finger, DHHC-type containing 12 |
| chr1_+_66820058 | 0.80 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr12_+_7053228 | 0.79 |
ENST00000540506.2
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr12_+_104680793 | 0.79 |
ENST00000529546.1
ENST00000529751.1 ENST00000540716.1 ENST00000528079.2 ENST00000526580.1 |
TXNRD1
|
thioredoxin reductase 1 |
| chr1_+_169079823 | 0.77 |
ENST00000367813.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr12_+_104680659 | 0.76 |
ENST00000526691.1
ENST00000531691.1 ENST00000388854.3 ENST00000354940.6 ENST00000526390.1 ENST00000531689.1 |
TXNRD1
|
thioredoxin reductase 1 |
| chr3_-_49058479 | 0.74 |
ENST00000440857.1
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr1_+_244816371 | 0.69 |
ENST00000263831.7
|
DESI2
|
desumoylating isopeptidase 2 |
| chr1_+_244214577 | 0.68 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr2_-_65659762 | 0.66 |
ENST00000440972.1
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr7_-_128415844 | 0.66 |
ENST00000249389.2
|
OPN1SW
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr2_+_128175997 | 0.65 |
ENST00000234071.3
ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr19_+_2476116 | 0.64 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr16_-_29875057 | 0.63 |
ENST00000219789.6
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr8_-_80993010 | 0.62 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr3_+_49058444 | 0.62 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr19_+_48325097 | 0.61 |
ENST00000221996.7
ENST00000539067.1 |
CRX
|
cone-rod homeobox |
| chr16_-_31076273 | 0.61 |
ENST00000426488.2
|
ZNF668
|
zinc finger protein 668 |
| chr17_+_12626199 | 0.61 |
ENST00000609971.1
|
AC005358.1
|
AC005358.1 |
| chr17_+_43318434 | 0.59 |
ENST00000587489.1
|
FMNL1
|
formin-like 1 |
| chr2_+_120770581 | 0.58 |
ENST00000263713.5
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr5_+_133451254 | 0.56 |
ENST00000517851.1
ENST00000521639.1 ENST00000522375.1 ENST00000378560.4 ENST00000432532.2 ENST00000520958.1 ENST00000518915.1 ENST00000395023.1 |
TCF7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr17_+_77020146 | 0.56 |
ENST00000579760.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr16_-_29874462 | 0.53 |
ENST00000566113.1
ENST00000569956.1 ENST00000570016.1 |
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr17_+_38465441 | 0.50 |
ENST00000577646.1
ENST00000254066.5 |
RARA
|
retinoic acid receptor, alpha |
| chr22_-_40929812 | 0.48 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr14_+_31046959 | 0.47 |
ENST00000547532.1
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr1_-_21606013 | 0.47 |
ENST00000357071.4
|
ECE1
|
endothelin converting enzyme 1 |
| chr14_-_23395623 | 0.46 |
ENST00000556043.1
|
PRMT5
|
protein arginine methyltransferase 5 |
| chr11_-_71752571 | 0.46 |
ENST00000544238.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chrX_+_153769409 | 0.46 |
ENST00000440286.1
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr11_-_71753188 | 0.46 |
ENST00000543009.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr14_+_52327350 | 0.46 |
ENST00000555472.1
ENST00000556766.1 |
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr14_-_101034811 | 0.45 |
ENST00000553553.1
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr9_-_130966497 | 0.43 |
ENST00000393608.1
ENST00000372948.3 |
CIZ1
|
CDKN1A interacting zinc finger protein 1 |
| chr3_-_19975665 | 0.43 |
ENST00000295824.9
ENST00000389256.4 |
EFHB
|
EF-hand domain family, member B |
| chr3_-_170626418 | 0.42 |
ENST00000474096.1
ENST00000295822.2 |
EIF5A2
|
eukaryotic translation initiation factor 5A2 |
| chr18_-_64271316 | 0.41 |
ENST00000540086.1
ENST00000580157.1 |
CDH19
|
cadherin 19, type 2 |
| chr9_-_39288092 | 0.40 |
ENST00000323947.7
ENST00000297668.6 ENST00000377656.2 ENST00000377659.1 |
CNTNAP3
|
contactin associated protein-like 3 |
| chr17_+_16945820 | 0.39 |
ENST00000577514.1
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr17_+_73521763 | 0.38 |
ENST00000167462.5
ENST00000375227.4 ENST00000392550.3 ENST00000578363.1 ENST00000579392.1 |
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr16_-_3074231 | 0.37 |
ENST00000572355.1
ENST00000248089.3 ENST00000574980.1 ENST00000354679.3 ENST00000396916.1 ENST00000573842.1 |
HCFC1R1
|
host cell factor C1 regulator 1 (XPO1 dependent) |
| chr2_-_27486951 | 0.36 |
ENST00000432351.1
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr9_+_20927764 | 0.36 |
ENST00000603044.1
ENST00000604254.1 |
FOCAD
|
focadhesin |
| chr17_+_77020224 | 0.35 |
ENST00000339142.2
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr11_-_71752838 | 0.35 |
ENST00000537930.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr2_+_189156389 | 0.34 |
ENST00000409843.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr7_-_97881429 | 0.34 |
ENST00000420697.1
ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr12_-_52604607 | 0.32 |
ENST00000551894.1
ENST00000553017.1 |
C12orf80
|
chromosome 12 open reading frame 80 |
| chr2_-_220252068 | 0.32 |
ENST00000430206.1
ENST00000429013.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr2_+_189156721 | 0.32 |
ENST00000409927.1
ENST00000409805.1 |
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr17_+_77020325 | 0.31 |
ENST00000311661.4
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr22_-_29663954 | 0.31 |
ENST00000216085.7
|
RHBDD3
|
rhomboid domain containing 3 |
| chr9_+_2015335 | 0.31 |
ENST00000349721.2
ENST00000357248.2 ENST00000450198.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_+_120770645 | 0.31 |
ENST00000443902.2
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr2_+_189156586 | 0.31 |
ENST00000409830.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr14_+_52327109 | 0.31 |
ENST00000335281.4
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chrX_+_17653413 | 0.30 |
ENST00000398097.3
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
| chr8_-_37457350 | 0.30 |
ENST00000519691.1
|
RP11-150O12.3
|
RP11-150O12.3 |
| chr12_-_12674032 | 0.29 |
ENST00000298573.4
|
DUSP16
|
dual specificity phosphatase 16 |
| chr1_-_26633480 | 0.29 |
ENST00000450041.1
|
UBXN11
|
UBX domain protein 11 |
| chr2_+_189156638 | 0.29 |
ENST00000410051.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr2_+_120517174 | 0.28 |
ENST00000263708.2
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr14_+_58894404 | 0.28 |
ENST00000554463.1
ENST00000555833.1 |
KIAA0586
|
KIAA0586 |
| chr18_-_64271363 | 0.28 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr11_-_67120974 | 0.27 |
ENST00000539074.1
ENST00000312419.3 |
POLD4
|
polymerase (DNA-directed), delta 4, accessory subunit |
| chr19_-_39340563 | 0.26 |
ENST00000601813.1
|
HNRNPL
|
heterogeneous nuclear ribonucleoprotein L |
| chr2_-_14541060 | 0.26 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr3_+_36421826 | 0.26 |
ENST00000273183.3
|
STAC
|
SH3 and cysteine rich domain |
| chr5_+_5140436 | 0.25 |
ENST00000511368.1
ENST00000274181.7 |
ADAMTS16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16 |
| chr2_+_120517717 | 0.25 |
ENST00000420482.1
ENST00000488279.2 |
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr16_+_87636474 | 0.24 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr13_+_36050881 | 0.24 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chrX_+_139791917 | 0.23 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chr19_-_49843539 | 0.22 |
ENST00000602554.1
ENST00000358234.4 |
CTC-301O7.4
|
CTC-301O7.4 |
| chr16_+_21623392 | 0.22 |
ENST00000562961.1
|
METTL9
|
methyltransferase like 9 |
| chr6_-_35480640 | 0.22 |
ENST00000428978.1
ENST00000322263.4 |
TULP1
|
tubby like protein 1 |
| chr17_+_68100989 | 0.22 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr1_+_114522049 | 0.21 |
ENST00000369551.1
ENST00000320334.4 |
OLFML3
|
olfactomedin-like 3 |
| chr16_-_69373396 | 0.21 |
ENST00000562595.1
ENST00000562081.1 ENST00000306875.4 |
COG8
|
component of oligomeric golgi complex 8 |
| chr9_+_17135016 | 0.21 |
ENST00000425824.1
ENST00000262360.5 ENST00000380641.4 |
CNTLN
|
centlein, centrosomal protein |
| chr1_+_244816237 | 0.21 |
ENST00000302550.11
|
DESI2
|
desumoylating isopeptidase 2 |
| chr15_+_54901540 | 0.21 |
ENST00000539562.2
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr9_-_35072585 | 0.20 |
ENST00000448530.1
|
VCP
|
valosin containing protein |
| chr12_-_53171128 | 0.19 |
ENST00000332411.2
|
KRT76
|
keratin 76 |
| chr14_+_32476072 | 0.18 |
ENST00000556949.1
|
RP11-187E13.2
|
Uncharacterized protein |
| chr14_-_71107921 | 0.17 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr10_-_35104185 | 0.17 |
ENST00000374789.3
ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3
|
par-3 family cell polarity regulator |
| chr17_+_68101117 | 0.17 |
ENST00000587698.1
ENST00000587892.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr2_-_152146385 | 0.16 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr1_+_18957500 | 0.16 |
ENST00000375375.3
|
PAX7
|
paired box 7 |
| chrX_-_21676442 | 0.16 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr10_-_76868931 | 0.16 |
ENST00000372700.3
ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13
|
dual specificity phosphatase 13 |
| chr9_-_13279563 | 0.15 |
ENST00000541718.1
|
MPDZ
|
multiple PDZ domain protein |
| chr19_+_40877583 | 0.15 |
ENST00000596470.1
|
PLD3
|
phospholipase D family, member 3 |
| chr1_-_11918988 | 0.15 |
ENST00000376468.3
|
NPPB
|
natriuretic peptide B |
| chr19_+_46001697 | 0.15 |
ENST00000451287.2
ENST00000324688.4 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr10_-_116418053 | 0.15 |
ENST00000277895.5
|
ABLIM1
|
actin binding LIM protein 1 |
| chr11_-_76155700 | 0.15 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr1_+_114447763 | 0.15 |
ENST00000369563.3
|
DCLRE1B
|
DNA cross-link repair 1B |
| chr4_+_103422499 | 0.15 |
ENST00000511926.1
ENST00000507079.1 |
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr9_+_116298778 | 0.14 |
ENST00000462143.1
|
RGS3
|
regulator of G-protein signaling 3 |
| chr18_-_70532906 | 0.14 |
ENST00000299430.2
ENST00000397929.1 |
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr7_-_29152509 | 0.14 |
ENST00000448959.1
|
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr12_+_53818855 | 0.13 |
ENST00000550839.1
|
AMHR2
|
anti-Mullerian hormone receptor, type II |
| chr14_+_21785693 | 0.13 |
ENST00000382933.4
ENST00000557351.1 |
RPGRIP1
|
retinitis pigmentosa GTPase regulator interacting protein 1 |
| chr9_-_13279589 | 0.13 |
ENST00000319217.7
|
MPDZ
|
multiple PDZ domain protein |
| chr10_-_126849626 | 0.12 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr10_-_76868866 | 0.12 |
ENST00000607487.1
|
DUSP13
|
dual specificity phosphatase 13 |
| chr7_+_138482695 | 0.12 |
ENST00000422794.2
ENST00000397602.3 ENST00000442682.2 ENST00000458494.1 ENST00000413208.1 |
TMEM213
|
transmembrane protein 213 |
| chr11_-_76155618 | 0.11 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr3_-_13028536 | 0.11 |
ENST00000450726.1
|
IQSEC1
|
IQ motif and Sec7 domain 1 |
| chr3_-_179692042 | 0.11 |
ENST00000468741.1
|
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr7_+_116660246 | 0.11 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr7_-_78400598 | 0.10 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr4_-_147442982 | 0.10 |
ENST00000511374.1
ENST00000264986.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr16_+_69373323 | 0.10 |
ENST00000254940.5
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr1_-_160313025 | 0.10 |
ENST00000368069.3
ENST00000241704.7 |
COPA
|
coatomer protein complex, subunit alpha |
| chr4_-_147442817 | 0.10 |
ENST00000507030.1
|
SLC10A7
|
solute carrier family 10, member 7 |
| chr3_-_112218205 | 0.10 |
ENST00000383680.4
|
BTLA
|
B and T lymphocyte associated |
| chr4_+_169552748 | 0.09 |
ENST00000504519.1
ENST00000512127.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr9_+_4839762 | 0.09 |
ENST00000448872.2
ENST00000441844.1 |
RCL1
|
RNA terminal phosphate cyclase-like 1 |
| chr12_-_13248562 | 0.09 |
ENST00000457134.2
ENST00000537302.1 |
GSG1
|
germ cell associated 1 |
| chr15_+_33603147 | 0.09 |
ENST00000415757.3
ENST00000389232.4 |
RYR3
|
ryanodine receptor 3 |
| chr17_-_4689727 | 0.09 |
ENST00000328739.5
ENST00000354194.4 |
VMO1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chr6_+_121756809 | 0.09 |
ENST00000282561.3
|
GJA1
|
gap junction protein, alpha 1, 43kDa |
| chrX_-_138724994 | 0.09 |
ENST00000536274.1
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr5_-_16738451 | 0.09 |
ENST00000274203.9
ENST00000515803.1 |
MYO10
|
myosin X |
| chr10_-_89577910 | 0.08 |
ENST00000308448.7
ENST00000541004.1 |
ATAD1
|
ATPase family, AAA domain containing 1 |
| chr11_-_8680383 | 0.08 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chrX_-_110655391 | 0.08 |
ENST00000356915.2
ENST00000356220.3 |
DCX
|
doublecortin |
| chr7_+_93535866 | 0.07 |
ENST00000429473.1
ENST00000430875.1 ENST00000428834.1 |
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr10_+_102222798 | 0.07 |
ENST00000343737.5
|
WNT8B
|
wingless-type MMTV integration site family, member 8B |
| chr11_+_61129827 | 0.07 |
ENST00000542946.1
|
TMEM138
|
transmembrane protein 138 |
| chr1_-_201140673 | 0.07 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr1_+_171227069 | 0.07 |
ENST00000354841.4
|
FMO1
|
flavin containing monooxygenase 1 |
| chr10_+_74451883 | 0.07 |
ENST00000373053.3
ENST00000357157.6 |
MCU
|
mitochondrial calcium uniporter |
| chrX_+_100878079 | 0.07 |
ENST00000471229.2
|
ARMCX3
|
armadillo repeat containing, X-linked 3 |
| chr11_-_40315640 | 0.07 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chrX_+_15767971 | 0.07 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr16_+_4421841 | 0.07 |
ENST00000304735.3
|
VASN
|
vasorin |
| chr14_-_65785502 | 0.06 |
ENST00000553754.1
|
CTD-2509G16.5
|
CTD-2509G16.5 |
| chr19_-_42931567 | 0.06 |
ENST00000244289.4
|
LIPE
|
lipase, hormone-sensitive |
| chr9_+_17134980 | 0.06 |
ENST00000380647.3
|
CNTLN
|
centlein, centrosomal protein |
| chr1_+_151682909 | 0.06 |
ENST00000326413.3
ENST00000442233.2 |
RIIAD1
AL589765.1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 Uncharacterized protein; cDNA FLJ36032 fis, clone TESTI2017069 |
| chrY_+_16634483 | 0.06 |
ENST00000382872.1
|
NLGN4Y
|
neuroligin 4, Y-linked |
| chr15_+_58430567 | 0.05 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr6_-_35480705 | 0.05 |
ENST00000229771.6
|
TULP1
|
tubby like protein 1 |
| chr1_+_36024107 | 0.05 |
ENST00000437806.1
|
NCDN
|
neurochondrin |
| chr12_-_13248705 | 0.05 |
ENST00000396310.2
|
GSG1
|
germ cell associated 1 |
| chrX_+_65382381 | 0.05 |
ENST00000519389.1
|
HEPH
|
hephaestin |
| chr15_+_58430368 | 0.05 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr7_+_93535817 | 0.05 |
ENST00000248572.5
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr17_+_44668035 | 0.05 |
ENST00000398238.4
ENST00000225282.8 |
NSF
|
N-ethylmaleimide-sensitive factor |
| chr1_+_162351503 | 0.05 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chrX_+_100333709 | 0.05 |
ENST00000372930.4
|
TMEM35
|
transmembrane protein 35 |
| chr1_-_26633067 | 0.04 |
ENST00000421827.2
ENST00000374215.1 ENST00000374223.1 ENST00000357089.4 ENST00000535108.1 ENST00000314675.7 ENST00000436301.2 ENST00000423664.1 ENST00000374221.3 |
UBXN11
|
UBX domain protein 11 |
| chr12_-_13248732 | 0.04 |
ENST00000396302.3
|
GSG1
|
germ cell associated 1 |
| chr3_-_179691866 | 0.04 |
ENST00000464614.1
ENST00000476138.1 ENST00000463761.1 |
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr12_-_14849470 | 0.04 |
ENST00000261170.3
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr10_-_94003003 | 0.03 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chrX_-_110655306 | 0.03 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr17_-_40021656 | 0.03 |
ENST00000319121.3
|
KLHL11
|
kelch-like family member 11 |
| chr10_-_74714533 | 0.03 |
ENST00000373032.3
|
PLA2G12B
|
phospholipase A2, group XIIB |
| chrX_+_135278908 | 0.03 |
ENST00000539015.1
ENST00000370683.1 |
FHL1
|
four and a half LIM domains 1 |
| chr8_+_55528627 | 0.03 |
ENST00000220676.1
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant) |
| chrX_+_65382433 | 0.03 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr1_+_11751748 | 0.03 |
ENST00000294485.5
|
DRAXIN
|
dorsal inhibitory axon guidance protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of GSC_GSC2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0043311 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.3 | 1.7 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.3 | 1.1 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.3 | 3.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.2 | 0.9 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.2 | 1.3 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.2 | 0.6 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.2 | 1.2 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 1.6 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 2.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 2.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 1.3 | GO:1902363 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 0.7 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.8 | GO:1903281 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 1.5 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 0.5 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.6 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 1.7 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.5 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.6 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 | 0.3 | GO:0045209 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.1 | 1.4 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 0.2 | GO:0072387 | flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.1 | 0.1 | GO:1902612 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.1 | 0.5 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.1 | 0.2 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.3 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.8 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.5 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.9 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.9 | GO:0070431 | nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 1.2 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 1.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 2.3 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.3 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 1.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 4.0 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.1 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) |
| 0.0 | 0.8 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.4 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.6 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:0031860 | telomeric loop formation(GO:0031627) telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 | 0.8 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 1.5 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 0.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 1.0 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.2 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.0 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.1 | 1.7 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 2.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.5 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 1.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.9 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 4.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 1.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 2.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.6 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 1.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.6 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.2 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.4 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.4 | 1.2 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.3 | 1.5 | GO:0098625 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.3 | 1.5 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.2 | 2.5 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 1.5 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 1.7 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.2 | 1.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 0.7 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 1.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 0.7 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.4 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.1 | 0.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 1.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 1.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 1.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.3 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 3.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 2.0 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.2 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.5 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 0.7 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 2.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 1.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 1.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.9 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |