Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
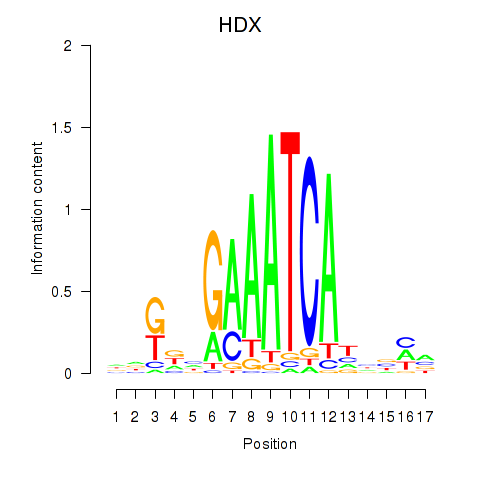
Results for HDX
Z-value: 0.49
Transcription factors associated with HDX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HDX
|
ENSG00000165259.9 | highly divergent homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HDX | hg19_v2_chrX_-_83757399_83757487 | 0.76 | 1.0e-04 | Click! |
Activity profile of HDX motif
Sorted Z-values of HDX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_113594279 | 3.91 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr1_-_153363452 | 3.62 |
ENST00000368732.1
ENST00000368733.3 |
S100A8
|
S100 calcium binding protein A8 |
| chr18_+_29027696 | 2.50 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr2_+_113735575 | 2.16 |
ENST00000376489.2
ENST00000259205.4 |
IL36G
|
interleukin 36, gamma |
| chr9_+_6215799 | 2.09 |
ENST00000417746.2
ENST00000456383.2 |
IL33
|
interleukin 33 |
| chr1_-_209825674 | 1.76 |
ENST00000367030.3
ENST00000356082.4 |
LAMB3
|
laminin, beta 3 |
| chr2_+_169926047 | 1.45 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr19_-_41934635 | 1.17 |
ENST00000321702.2
|
B3GNT8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
| chr19_-_58485895 | 0.96 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr3_-_51533966 | 0.82 |
ENST00000504652.1
|
VPRBP
|
Vpr (HIV-1) binding protein |
| chr8_+_24151620 | 0.79 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr14_+_71788096 | 0.64 |
ENST00000557151.1
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr3_+_107241882 | 0.64 |
ENST00000416476.2
|
BBX
|
bobby sox homolog (Drosophila) |
| chr1_+_47489240 | 0.60 |
ENST00000371901.3
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1 |
| chr16_+_1877204 | 0.37 |
ENST00000427358.2
|
FAHD1
|
fumarylacetoacetate hydrolase domain containing 1 |
| chr10_-_14372870 | 0.36 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chr13_+_97928395 | 0.33 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr6_+_42584847 | 0.31 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr12_+_10460417 | 0.23 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr11_+_9685604 | 0.18 |
ENST00000447399.2
ENST00000318950.6 |
SWAP70
|
SWAP switching B-cell complex 70kDa subunit |
| chr10_+_45406627 | 0.17 |
ENST00000389583.4
|
TMEM72
|
transmembrane protein 72 |
| chr8_+_145065705 | 0.16 |
ENST00000533044.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr12_-_118797475 | 0.12 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr7_+_144015218 | 0.11 |
ENST00000408951.1
|
OR2A1
|
olfactory receptor, family 2, subfamily A, member 1 |
| chr1_-_222014008 | 0.10 |
ENST00000431729.1
|
RP11-191N8.2
|
RP11-191N8.2 |
| chr22_+_40440804 | 0.09 |
ENST00000441751.1
ENST00000301923.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr3_-_100565249 | 0.05 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr22_+_45714672 | 0.03 |
ENST00000424557.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr1_+_117602925 | 0.03 |
ENST00000369466.4
|
TTF2
|
transcription termination factor, RNA polymerase II |
| chr5_+_157602404 | 0.02 |
ENST00000522975.1
|
CTC-436K13.1
|
CTC-436K13.1 |
| chrX_+_27608490 | 0.01 |
ENST00000451261.2
|
DCAF8L2
|
DDB1 and CUL4 associated factor 8-like 2 |
| chr1_+_84629976 | 0.01 |
ENST00000446538.1
ENST00000370684.1 ENST00000436133.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr11_-_66675371 | 0.00 |
ENST00000393955.2
|
PC
|
pyruvate carboxylase |
Network of associatons between targets according to the STRING database.
First level regulatory network of HDX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.9 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.4 | 2.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.2 | 1.2 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 3.6 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.1 | 1.5 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.2 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 1.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 2.5 | GO:0070268 | cornification(GO:0070268) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 2.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 3.6 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.6 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 6.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 1.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 1.5 | GO:0047035 | alcohol dehydrogenase (NAD) activity(GO:0004022) testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 0.4 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.6 | GO:0070330 | aromatase activity(GO:0070330) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 3.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 3.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.8 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |