Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
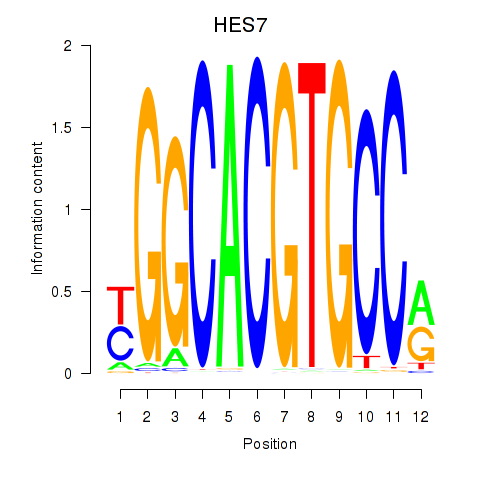
Results for HES7_HES5
Z-value: 0.64
Transcription factors associated with HES7_HES5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HES7
|
ENSG00000179111.4 | hes family bHLH transcription factor 7 |
|
HES5
|
ENSG00000197921.5 | hes family bHLH transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HES5 | hg19_v2_chr1_-_2461684_2461710 | 0.69 | 7.4e-04 | Click! |
| HES7 | hg19_v2_chr17_-_8027402_8027432 | -0.53 | 1.6e-02 | Click! |
Activity profile of HES7_HES5 motif
Sorted Z-values of HES7_HES5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_48326764 | 3.13 |
ENST00000413668.1
ENST00000441948.1 |
SLC38A5
|
solute carrier family 38, member 5 |
| chr13_+_43597269 | 2.92 |
ENST00000379221.2
|
DNAJC15
|
DnaJ (Hsp40) homolog, subfamily C, member 15 |
| chrX_-_48326683 | 2.39 |
ENST00000440085.1
|
SLC38A5
|
solute carrier family 38, member 5 |
| chr2_+_27665232 | 1.69 |
ENST00000543753.1
ENST00000288873.3 |
KRTCAP3
|
keratinocyte associated protein 3 |
| chr19_+_13106383 | 1.43 |
ENST00000397661.2
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr2_+_27665289 | 1.40 |
ENST00000407293.1
|
KRTCAP3
|
keratinocyte associated protein 3 |
| chr3_-_69062764 | 1.37 |
ENST00000295571.5
|
EOGT
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr3_-_69062790 | 1.34 |
ENST00000540955.1
ENST00000456376.1 |
EOGT
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr20_+_45523227 | 1.26 |
ENST00000327619.5
ENST00000357410.3 |
EYA2
|
eyes absent homolog 2 (Drosophila) |
| chr19_-_19739007 | 1.22 |
ENST00000586703.1
ENST00000591042.1 ENST00000407877.3 |
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr3_-_69062742 | 1.14 |
ENST00000424374.1
|
EOGT
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr1_+_16083154 | 1.12 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr2_-_232328867 | 1.06 |
ENST00000453992.1
ENST00000417652.1 ENST00000454824.1 |
NCL
|
nucleolin |
| chr19_+_45281118 | 1.00 |
ENST00000270279.3
ENST00000341505.4 |
CBLC
|
Cbl proto-oncogene C, E3 ubiquitin protein ligase |
| chr1_+_150254936 | 0.96 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr21_-_33985127 | 0.89 |
ENST00000290155.3
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr22_+_41777927 | 0.84 |
ENST00000266304.4
|
TEF
|
thyrotrophic embryonic factor |
| chr19_-_36822551 | 0.76 |
ENST00000591372.1
|
LINC00665
|
long intergenic non-protein coding RNA 665 |
| chr19_+_58095501 | 0.73 |
ENST00000536878.2
ENST00000597850.1 ENST00000597219.1 ENST00000598689.1 ENST00000599456.1 ENST00000307468.4 |
ZIK1
|
zinc finger protein interacting with K protein 1 |
| chr9_+_706842 | 0.67 |
ENST00000382293.3
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr17_+_30813576 | 0.62 |
ENST00000313401.3
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr5_+_112227311 | 0.60 |
ENST00000391338.1
|
ZRSR1
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 1 |
| chr11_+_45944190 | 0.60 |
ENST00000401752.1
ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr6_+_46620676 | 0.59 |
ENST00000371347.5
ENST00000411689.2 |
SLC25A27
|
solute carrier family 25, member 27 |
| chr4_+_2965307 | 0.59 |
ENST00000398051.4
ENST00000503518.2 ENST00000398052.4 ENST00000345167.6 ENST00000504933.1 ENST00000442472.2 |
GRK4
|
G protein-coupled receptor kinase 4 |
| chr8_+_38089198 | 0.58 |
ENST00000528358.1
ENST00000529642.1 ENST00000532222.1 ENST00000520272.2 |
DDHD2
|
DDHD domain containing 2 |
| chr3_-_149510553 | 0.57 |
ENST00000462519.2
ENST00000446160.1 ENST00000383050.3 |
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr20_-_44539538 | 0.56 |
ENST00000372420.1
|
PLTP
|
phospholipid transfer protein |
| chr3_+_39851170 | 0.56 |
ENST00000425621.1
ENST00000396217.3 |
MYRIP
|
myosin VIIA and Rab interacting protein |
| chr20_-_45985464 | 0.56 |
ENST00000458360.2
ENST00000262975.4 |
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr6_-_112194484 | 0.55 |
ENST00000518295.1
ENST00000484067.2 ENST00000229470.5 ENST00000356013.2 ENST00000368678.4 ENST00000523238.1 ENST00000354650.3 |
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr4_+_2819883 | 0.55 |
ENST00000511747.1
ENST00000503393.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr2_-_219154715 | 0.54 |
ENST00000451181.1
ENST00000429501.1 |
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chrX_+_105969893 | 0.52 |
ENST00000255499.2
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr5_+_148521381 | 0.52 |
ENST00000504238.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr20_-_45985414 | 0.52 |
ENST00000461685.1
ENST00000372023.3 ENST00000540497.1 ENST00000435836.1 ENST00000471951.2 ENST00000352431.2 ENST00000396281.4 ENST00000355972.4 ENST00000360911.3 |
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr2_+_201171268 | 0.50 |
ENST00000423749.1
ENST00000428692.1 ENST00000457757.1 ENST00000453663.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr20_+_35201993 | 0.50 |
ENST00000373872.4
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr19_-_19739321 | 0.49 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr14_+_24641062 | 0.49 |
ENST00000311457.3
ENST00000557806.1 ENST00000559919.1 |
REC8
|
REC8 meiotic recombination protein |
| chr6_+_13182751 | 0.48 |
ENST00000415087.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr4_-_53522703 | 0.47 |
ENST00000508499.1
|
USP46
|
ubiquitin specific peptidase 46 |
| chr4_+_184427235 | 0.46 |
ENST00000412117.1
ENST00000434682.2 |
ING2
|
inhibitor of growth family, member 2 |
| chr16_-_21416640 | 0.46 |
ENST00000542817.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr5_+_78532003 | 0.44 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr19_-_53632925 | 0.42 |
ENST00000595174.1
|
ZNF415
|
zinc finger protein 415 |
| chr1_+_210111534 | 0.41 |
ENST00000422431.1
ENST00000534859.1 ENST00000399639.2 ENST00000537238.1 |
SYT14
|
synaptotagmin XIV |
| chr1_+_210111570 | 0.41 |
ENST00000367019.1
ENST00000472886.1 |
SYT14
|
synaptotagmin XIV |
| chr11_+_134146635 | 0.41 |
ENST00000431683.2
|
GLB1L3
|
galactosidase, beta 1-like 3 |
| chr2_-_237123244 | 0.41 |
ENST00000447030.1
|
ASB18
|
ankyrin repeat and SOCS box containing 18 |
| chr1_-_35497506 | 0.41 |
ENST00000317538.5
ENST00000373340.2 ENST00000357182.4 |
ZMYM6
|
zinc finger, MYM-type 6 |
| chr5_+_179125907 | 0.41 |
ENST00000247461.4
ENST00000452673.2 ENST00000502498.1 ENST00000507307.1 ENST00000513246.1 ENST00000502673.1 ENST00000506654.1 ENST00000512607.2 ENST00000510810.1 |
CANX
|
calnexin |
| chr6_+_46620705 | 0.39 |
ENST00000452689.2
|
SLC25A27
|
solute carrier family 25, member 27 |
| chr2_+_201171064 | 0.39 |
ENST00000451764.2
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr5_+_148521136 | 0.38 |
ENST00000506113.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr6_-_114292449 | 0.38 |
ENST00000519065.1
|
HDAC2
|
histone deacetylase 2 |
| chr5_+_148521046 | 0.38 |
ENST00000326685.7
ENST00000356541.3 ENST00000309868.7 |
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr3_+_39851094 | 0.38 |
ENST00000302541.6
|
MYRIP
|
myosin VIIA and Rab interacting protein |
| chr3_-_189839467 | 0.37 |
ENST00000426003.1
|
LEPREL1
|
leprecan-like 1 |
| chr20_+_35201857 | 0.36 |
ENST00000373874.2
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr1_+_45140400 | 0.36 |
ENST00000453711.1
|
C1orf228
|
chromosome 1 open reading frame 228 |
| chr11_-_116968987 | 0.35 |
ENST00000434315.2
ENST00000292055.4 ENST00000375288.1 ENST00000542607.1 ENST00000445177.1 ENST00000375300.1 ENST00000446921.2 |
SIK3
|
SIK family kinase 3 |
| chr6_-_143771799 | 0.35 |
ENST00000237283.8
|
ADAT2
|
adenosine deaminase, tRNA-specific 2 |
| chr13_+_115047097 | 0.35 |
ENST00000351487.5
|
UPF3A
|
UPF3 regulator of nonsense transcripts homolog A (yeast) |
| chr7_+_5468362 | 0.34 |
ENST00000608012.1
|
RP11-1275H24.3
|
RP11-1275H24.3 |
| chr10_+_131265443 | 0.34 |
ENST00000306010.7
|
MGMT
|
O-6-methylguanine-DNA methyltransferase |
| chr14_+_104689918 | 0.34 |
ENST00000553757.1
ENST00000556528.1 ENST00000555282.1 |
RP11-260M19.2
|
RP11-260M19.2 |
| chr14_-_36990061 | 0.33 |
ENST00000546983.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr22_-_30752861 | 0.33 |
ENST00000439242.1
ENST00000215793.8 |
SF3A1
|
splicing factor 3a, subunit 1, 120kDa |
| chr2_+_201170770 | 0.32 |
ENST00000409988.3
ENST00000409385.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chrX_+_134124968 | 0.32 |
ENST00000330288.4
|
SMIM10
|
small integral membrane protein 10 |
| chr16_-_21849091 | 0.32 |
ENST00000537951.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr4_+_94750014 | 0.31 |
ENST00000306011.3
|
ATOH1
|
atonal homolog 1 (Drosophila) |
| chr14_-_77737543 | 0.31 |
ENST00000298352.4
|
NGB
|
neuroglobin |
| chr5_+_148521454 | 0.30 |
ENST00000508983.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr2_+_201170596 | 0.30 |
ENST00000439084.1
ENST00000409718.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr22_-_43583079 | 0.29 |
ENST00000216129.6
|
TTLL12
|
tubulin tyrosine ligase-like family, member 12 |
| chr6_-_114292284 | 0.29 |
ENST00000520895.1
ENST00000521163.1 ENST00000524334.1 ENST00000368632.2 ENST00000398283.2 |
HDAC2
|
histone deacetylase 2 |
| chr16_-_23160591 | 0.29 |
ENST00000219689.7
|
USP31
|
ubiquitin specific peptidase 31 |
| chr12_-_76953513 | 0.29 |
ENST00000547540.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr4_+_113152881 | 0.29 |
ENST00000274000.5
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr7_+_148959262 | 0.28 |
ENST00000434415.1
|
ZNF783
|
zinc finger family member 783 |
| chr22_-_42310570 | 0.28 |
ENST00000457093.1
|
SHISA8
|
shisa family member 8 |
| chr5_+_95066823 | 0.28 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr15_+_81489213 | 0.28 |
ENST00000559383.1
ENST00000394660.2 |
IL16
|
interleukin 16 |
| chr21_+_42694732 | 0.27 |
ENST00000398646.3
|
FAM3B
|
family with sequence similarity 3, member B |
| chr16_-_30237150 | 0.27 |
ENST00000543463.1
|
RP11-347C12.1
|
Putative NPIP-like protein LOC613037 |
| chr13_+_80055284 | 0.27 |
ENST00000218652.7
|
NDFIP2
|
Nedd4 family interacting protein 2 |
| chr18_+_11752040 | 0.27 |
ENST00000423027.3
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr1_-_45140227 | 0.26 |
ENST00000372237.3
|
TMEM53
|
transmembrane protein 53 |
| chr19_+_37837185 | 0.25 |
ENST00000541583.2
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr19_+_4304685 | 0.25 |
ENST00000601006.1
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr6_-_30712313 | 0.24 |
ENST00000376377.2
ENST00000259874.5 |
IER3
|
immediate early response 3 |
| chr13_+_80055581 | 0.24 |
ENST00000487865.1
|
NDFIP2
|
Nedd4 family interacting protein 2 |
| chr2_-_132919528 | 0.24 |
ENST00000409867.1
|
ANKRD30BL
|
ankyrin repeat domain 30B-like |
| chr4_+_76439665 | 0.23 |
ENST00000508105.1
ENST00000311638.3 ENST00000380837.3 ENST00000507556.1 ENST00000504190.1 ENST00000507885.1 ENST00000502620.1 ENST00000514480.1 |
THAP6
|
THAP domain containing 6 |
| chr22_+_42095497 | 0.23 |
ENST00000401548.3
ENST00000540833.1 ENST00000400107.1 ENST00000300398.4 |
MEI1
|
meiosis inhibitor 1 |
| chr1_+_44457441 | 0.22 |
ENST00000466180.1
|
CCDC24
|
coiled-coil domain containing 24 |
| chr11_-_14380664 | 0.22 |
ENST00000545643.1
ENST00000256196.4 |
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr2_+_201170703 | 0.22 |
ENST00000358677.5
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr8_+_104383759 | 0.22 |
ENST00000415886.2
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr14_+_77647966 | 0.22 |
ENST00000554766.1
|
TMEM63C
|
transmembrane protein 63C |
| chr5_-_114598548 | 0.22 |
ENST00000379615.3
ENST00000419445.1 |
PGGT1B
|
protein geranylgeranyltransferase type I, beta subunit |
| chr2_+_201171372 | 0.22 |
ENST00000409140.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr9_+_92219919 | 0.21 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr1_+_44445549 | 0.21 |
ENST00000356836.6
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr2_+_201171242 | 0.21 |
ENST00000360760.5
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr4_+_76439649 | 0.21 |
ENST00000507557.1
|
THAP6
|
THAP domain containing 6 |
| chr21_+_45432174 | 0.21 |
ENST00000380221.3
ENST00000291574.4 |
TRAPPC10
|
trafficking protein particle complex 10 |
| chr21_-_46330545 | 0.21 |
ENST00000320216.6
ENST00000397852.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr22_+_18560675 | 0.21 |
ENST00000329627.7
|
PEX26
|
peroxisomal biogenesis factor 26 |
| chr19_-_4723761 | 0.20 |
ENST00000597849.1
ENST00000598800.1 ENST00000602161.1 ENST00000597726.1 ENST00000601130.1 ENST00000262960.9 |
DPP9
|
dipeptidyl-peptidase 9 |
| chr17_+_39344697 | 0.20 |
ENST00000377723.3
|
KRTAP9-1
|
keratin associated protein 9-1 |
| chr5_-_143550241 | 0.20 |
ENST00000522203.1
|
YIPF5
|
Yip1 domain family, member 5 |
| chr12_-_76953453 | 0.19 |
ENST00000549570.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr1_+_16083098 | 0.19 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr12_+_6644443 | 0.19 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr4_+_188916918 | 0.19 |
ENST00000509524.1
ENST00000326866.4 |
ZFP42
|
ZFP42 zinc finger protein |
| chr20_-_20693131 | 0.18 |
ENST00000202677.7
|
RALGAPA2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr8_+_104383728 | 0.18 |
ENST00000330295.5
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr4_+_1003742 | 0.17 |
ENST00000398484.2
|
FGFRL1
|
fibroblast growth factor receptor-like 1 |
| chr14_-_67826486 | 0.17 |
ENST00000555431.1
ENST00000554236.1 ENST00000555474.1 |
ATP6V1D
|
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
| chr14_-_81687197 | 0.17 |
ENST00000553612.1
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa |
| chr9_-_23821842 | 0.17 |
ENST00000544538.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr17_-_1531635 | 0.17 |
ENST00000571650.1
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr18_-_46987000 | 0.16 |
ENST00000442713.2
ENST00000269445.6 |
DYM
|
dymeclin |
| chr3_+_38080691 | 0.16 |
ENST00000308059.6
ENST00000346219.3 ENST00000452631.2 |
DLEC1
|
deleted in lung and esophageal cancer 1 |
| chr12_-_76953573 | 0.16 |
ENST00000549646.1
ENST00000550628.1 ENST00000553139.1 ENST00000261183.3 ENST00000393250.4 |
OSBPL8
|
oxysterol binding protein-like 8 |
| chr14_-_51297197 | 0.16 |
ENST00000382043.4
|
NIN
|
ninein (GSK3B interacting protein) |
| chr15_-_43212836 | 0.15 |
ENST00000566931.1
ENST00000564431.1 ENST00000567274.1 |
TTBK2
|
tau tubulin kinase 2 |
| chr13_+_115079949 | 0.15 |
ENST00000361283.1
|
CHAMP1
|
chromosome alignment maintaining phosphoprotein 1 |
| chr12_+_25205155 | 0.15 |
ENST00000550945.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr5_+_143550396 | 0.15 |
ENST00000512467.1
|
KCTD16
|
potassium channel tetramerization domain containing 16 |
| chrX_-_153523462 | 0.14 |
ENST00000361930.3
ENST00000369926.1 |
TEX28
|
testis expressed 28 |
| chr17_+_5185552 | 0.14 |
ENST00000262477.6
ENST00000408982.2 ENST00000575991.1 ENST00000537505.1 ENST00000546142.2 |
RABEP1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr2_+_201170999 | 0.14 |
ENST00000439395.1
ENST00000444012.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr16_+_222846 | 0.14 |
ENST00000251595.6
ENST00000397806.1 |
HBA2
|
hemoglobin, alpha 2 |
| chr2_-_220118631 | 0.14 |
ENST00000248437.4
|
TUBA4A
|
tubulin, alpha 4a |
| chr2_+_149402009 | 0.13 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chrX_+_152224766 | 0.13 |
ENST00000370265.4
ENST00000447306.1 |
PNMA3
|
paraneoplastic Ma antigen 3 |
| chr3_+_52719936 | 0.13 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr14_+_67826709 | 0.13 |
ENST00000256383.4
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr11_-_26593649 | 0.13 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr22_-_30987837 | 0.12 |
ENST00000335214.6
|
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chr19_-_19049791 | 0.12 |
ENST00000594439.1
ENST00000221222.11 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr11_-_26593677 | 0.12 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr7_-_6570817 | 0.12 |
ENST00000435185.1
|
GRID2IP
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein |
| chr1_-_220220000 | 0.12 |
ENST00000366923.3
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr14_-_51297360 | 0.11 |
ENST00000496749.1
|
NIN
|
ninein (GSK3B interacting protein) |
| chr4_-_52904425 | 0.11 |
ENST00000535450.1
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr10_-_71169031 | 0.11 |
ENST00000373307.1
|
TACR2
|
tachykinin receptor 2 |
| chr11_+_64059464 | 0.11 |
ENST00000394525.2
|
KCNK4
|
potassium channel, subfamily K, member 4 |
| chr4_-_17783135 | 0.11 |
ENST00000265018.3
|
FAM184B
|
family with sequence similarity 184, member B |
| chr1_-_220219775 | 0.11 |
ENST00000609181.1
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr7_+_101460882 | 0.11 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr10_+_72194585 | 0.10 |
ENST00000420338.2
|
AC022532.1
|
Uncharacterized protein |
| chr9_-_139658965 | 0.10 |
ENST00000316144.5
|
LCN15
|
lipocalin 15 |
| chr15_-_65477637 | 0.10 |
ENST00000300107.3
|
CLPX
|
caseinolytic mitochondrial matrix peptidase chaperone subunit |
| chr1_-_109655377 | 0.10 |
ENST00000369948.3
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr22_-_30987849 | 0.10 |
ENST00000402284.3
ENST00000354694.7 |
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chr1_-_231114542 | 0.10 |
ENST00000522821.1
ENST00000366661.4 ENST00000366662.4 ENST00000414259.1 ENST00000522399.1 |
TTC13
|
tetratricopeptide repeat domain 13 |
| chr5_+_115298165 | 0.10 |
ENST00000357872.4
|
AQPEP
|
Aminopeptidase Q |
| chr9_+_131451480 | 0.09 |
ENST00000322030.8
|
SET
|
SET nuclear oncogene |
| chr1_+_44445643 | 0.09 |
ENST00000309519.7
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr2_+_113403434 | 0.09 |
ENST00000272542.3
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1 |
| chr12_-_6756559 | 0.09 |
ENST00000536350.1
ENST00000414226.2 ENST00000546114.1 |
ACRBP
|
acrosin binding protein |
| chr5_+_152870215 | 0.09 |
ENST00000518142.1
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1 |
| chr19_-_55954230 | 0.09 |
ENST00000376325.4
|
SHISA7
|
shisa family member 7 |
| chr12_-_6756609 | 0.09 |
ENST00000229243.2
|
ACRBP
|
acrosin binding protein |
| chr12_-_104350898 | 0.09 |
ENST00000552940.1
ENST00000547975.1 ENST00000549478.1 ENST00000546540.1 ENST00000546819.1 ENST00000378090.4 ENST00000547945.1 |
C12orf73
|
chromosome 12 open reading frame 73 |
| chr15_-_57025728 | 0.08 |
ENST00000559352.1
|
ZNF280D
|
zinc finger protein 280D |
| chr1_-_93426998 | 0.08 |
ENST00000370310.4
|
FAM69A
|
family with sequence similarity 69, member A |
| chr8_-_142012169 | 0.08 |
ENST00000517453.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr1_+_28832455 | 0.08 |
ENST00000398958.2
ENST00000427469.1 ENST00000434290.1 ENST00000373833.6 |
RCC1
|
regulator of chromosome condensation 1 |
| chr17_+_48172639 | 0.08 |
ENST00000503176.1
ENST00000503614.1 |
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2 |
| chr1_+_150293973 | 0.07 |
ENST00000414970.2
ENST00000543398.1 |
PRPF3
|
pre-mRNA processing factor 3 |
| chr10_+_180405 | 0.07 |
ENST00000439456.1
ENST00000397962.3 ENST00000309776.4 ENST00000381602.4 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr2_+_65216462 | 0.07 |
ENST00000234256.3
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr4_+_184580420 | 0.07 |
ENST00000334690.6
ENST00000357207.4 |
TRAPPC11
|
trafficking protein particle complex 11 |
| chr1_+_150293921 | 0.07 |
ENST00000324862.6
|
PRPF3
|
pre-mRNA processing factor 3 |
| chr19_-_55458860 | 0.07 |
ENST00000592784.1
ENST00000448121.2 ENST00000340844.2 |
NLRP7
|
NLR family, pyrin domain containing 7 |
| chr11_+_35684288 | 0.07 |
ENST00000299413.5
|
TRIM44
|
tripartite motif containing 44 |
| chr1_+_44457261 | 0.07 |
ENST00000372318.3
|
CCDC24
|
coiled-coil domain containing 24 |
| chr14_+_39735411 | 0.07 |
ENST00000603904.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr21_-_47575481 | 0.07 |
ENST00000291670.5
ENST00000397748.1 ENST00000359679.2 ENST00000355384.2 ENST00000397746.3 ENST00000397743.1 |
FTCD
|
formimidoyltransferase cyclodeaminase |
| chr1_-_1293904 | 0.06 |
ENST00000309212.6
ENST00000342753.4 ENST00000445648.2 |
MXRA8
|
matrix-remodelling associated 8 |
| chr7_-_53879623 | 0.06 |
ENST00000380970.2
|
GS1-179L18.1
|
GS1-179L18.1 |
| chr22_+_24891210 | 0.06 |
ENST00000382760.2
|
UPB1
|
ureidopropionase, beta |
| chr5_+_115298145 | 0.06 |
ENST00000395528.2
|
AQPEP
|
Aminopeptidase Q |
| chr22_-_24181174 | 0.06 |
ENST00000318109.7
ENST00000406855.3 ENST00000404056.1 ENST00000476077.1 |
DERL3
|
derlin 3 |
| chr12_+_52345448 | 0.06 |
ENST00000257963.4
ENST00000541224.1 ENST00000426655.2 ENST00000536420.1 ENST00000415850.2 |
ACVR1B
|
activin A receptor, type IB |
| chr5_+_152870106 | 0.06 |
ENST00000285900.5
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1 |
| chr5_-_143550159 | 0.06 |
ENST00000448443.2
ENST00000513112.1 ENST00000519064.1 ENST00000274496.5 |
YIPF5
|
Yip1 domain family, member 5 |
| chr21_-_16374688 | 0.06 |
ENST00000411932.1
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr4_-_6692246 | 0.06 |
ENST00000499502.2
|
RP11-539L10.2
|
RP11-539L10.2 |
| chr4_+_76439095 | 0.06 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr22_-_25170683 | 0.06 |
ENST00000332271.5
|
PIWIL3
|
piwi-like RNA-mediated gene silencing 3 |
| chrX_+_16804544 | 0.06 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr22_-_50523760 | 0.05 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr17_+_27046988 | 0.05 |
ENST00000496182.1
|
RPL23A
|
ribosomal protein L23a |
Network of associatons between targets according to the STRING database.
First level regulatory network of HES7_HES5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.9 | GO:1902957 | negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 0.3 | 1.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.3 | 5.5 | GO:0015816 | glycine transport(GO:0015816) |
| 0.2 | 0.7 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.2 | 0.6 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 1.1 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 0.6 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.1 | 0.3 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.1 | 0.3 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.1 | 0.7 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.1 | 0.3 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.1 | 0.5 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) |
| 0.1 | 0.2 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.1 | 1.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 0.2 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 | 0.6 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 0.9 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 0.4 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.5 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 1.0 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.8 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 1.7 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.6 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.5 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.0 | 0.9 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.3 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.6 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.0 | 0.4 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.3 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 4.0 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.6 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.4 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.2 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.2 | GO:0045141 | meiotic telomere clustering(GO:0045141) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 1.6 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.3 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.6 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.5 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.2 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.1 | 0.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) glial limiting end-foot(GO:0097451) |
| 0.0 | 0.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.6 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 2.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.5 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:0071004 | U2 snRNP(GO:0005686) U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 1.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.8 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.4 | 5.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 0.6 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.2 | 0.6 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.2 | 1.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 1.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.3 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 0.3 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.2 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.1 | 0.2 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 0.6 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.6 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 1.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.4 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 1.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.2 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.7 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 1.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.3 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.6 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |