Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for HINFP
Z-value: 0.91
Transcription factors associated with HINFP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HINFP
|
ENSG00000172273.8 | histone H4 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HINFP | hg19_v2_chr11_+_118992269_118992334 | -0.09 | 7.1e-01 | Click! |
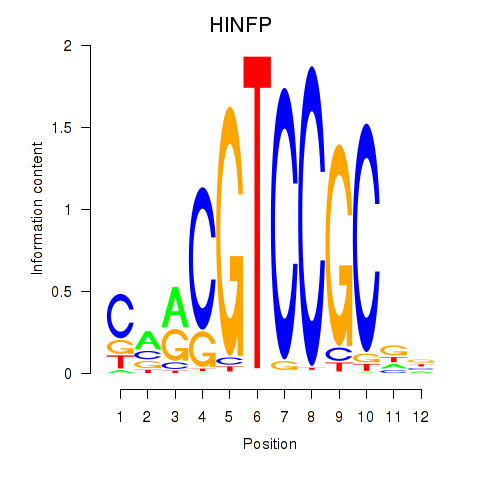
Activity profile of HINFP motif
Sorted Z-values of HINFP motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_54963026 | 2.67 |
ENST00000560208.1
ENST00000557792.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr8_+_32405728 | 2.63 |
ENST00000523079.1
ENST00000338921.4 ENST00000356819.4 ENST00000287845.5 ENST00000341377.5 |
NRG1
|
neuregulin 1 |
| chr9_+_130965651 | 2.39 |
ENST00000475805.1
ENST00000341179.7 ENST00000372923.3 |
DNM1
|
dynamin 1 |
| chr1_-_197115818 | 2.17 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr8_+_32405785 | 2.13 |
ENST00000287842.3
|
NRG1
|
neuregulin 1 |
| chr16_-_54962704 | 2.10 |
ENST00000502066.2
ENST00000560912.1 ENST00000558952.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr9_+_130965677 | 2.10 |
ENST00000393594.3
ENST00000486160.1 |
DNM1
|
dynamin 1 |
| chr12_-_90103077 | 1.90 |
ENST00000551310.1
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr8_+_32406137 | 1.87 |
ENST00000521670.1
|
NRG1
|
neuregulin 1 |
| chr12_+_65672423 | 1.84 |
ENST00000355192.3
ENST00000308259.5 ENST00000540804.1 ENST00000535664.1 ENST00000541189.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr2_+_10442993 | 1.76 |
ENST00000423674.1
ENST00000307845.3 |
HPCAL1
|
hippocalcin-like 1 |
| chr8_-_27462822 | 1.73 |
ENST00000522098.1
|
CLU
|
clusterin |
| chr11_+_62104897 | 1.68 |
ENST00000415229.2
ENST00000535727.1 ENST00000301776.5 |
ASRGL1
|
asparaginase like 1 |
| chr5_+_72143988 | 1.54 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr8_+_32406179 | 1.53 |
ENST00000405005.3
|
NRG1
|
neuregulin 1 |
| chr16_+_2510081 | 1.52 |
ENST00000361837.4
ENST00000569496.1 ENST00000567489.1 ENST00000563531.1 ENST00000483320.1 |
C16orf59
|
chromosome 16 open reading frame 59 |
| chr9_+_137533615 | 1.52 |
ENST00000371817.3
|
COL5A1
|
collagen, type V, alpha 1 |
| chr17_+_17206635 | 1.50 |
ENST00000389022.4
|
NT5M
|
5',3'-nucleotidase, mitochondrial |
| chr3_+_110790590 | 1.47 |
ENST00000485303.1
|
PVRL3
|
poliovirus receptor-related 3 |
| chr10_+_94352956 | 1.47 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr19_-_55919087 | 1.44 |
ENST00000587845.1
ENST00000589978.1 ENST00000264552.9 |
UBE2S
|
ubiquitin-conjugating enzyme E2S |
| chr12_+_65672702 | 1.37 |
ENST00000538045.1
ENST00000535239.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr17_-_46690839 | 1.37 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr17_+_29158962 | 1.29 |
ENST00000321990.4
|
ATAD5
|
ATPase family, AAA domain containing 5 |
| chr12_-_90102594 | 1.28 |
ENST00000428670.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr2_+_30670127 | 1.28 |
ENST00000540623.1
ENST00000476038.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr19_+_36545833 | 1.27 |
ENST00000401500.2
ENST00000270301.7 ENST00000427823.2 |
WDR62
|
WD repeat domain 62 |
| chr4_+_123747834 | 1.26 |
ENST00000264498.3
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr7_-_134143841 | 1.25 |
ENST00000285930.4
|
AKR1B1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr6_+_159291090 | 1.24 |
ENST00000367073.4
ENST00000608817.1 |
C6orf99
|
chromosome 6 open reading frame 99 |
| chr3_+_110790715 | 1.23 |
ENST00000319792.3
|
PVRL3
|
poliovirus receptor-related 3 |
| chrX_+_21392529 | 1.22 |
ENST00000425654.2
ENST00000543067.1 |
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr11_-_75062730 | 1.22 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr1_+_109102652 | 1.20 |
ENST00000370035.3
ENST00000405454.1 |
FAM102B
|
family with sequence similarity 102, member B |
| chr17_+_48450575 | 1.15 |
ENST00000338165.4
ENST00000393271.2 ENST00000511519.2 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
| chr4_+_123747979 | 1.12 |
ENST00000608478.1
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr11_-_6440283 | 1.11 |
ENST00000299402.6
ENST00000609360.1 ENST00000389906.2 ENST00000532020.2 |
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr3_+_110790867 | 1.10 |
ENST00000486596.1
ENST00000493615.1 |
PVRL3
|
poliovirus receptor-related 3 |
| chr19_+_36545781 | 1.08 |
ENST00000388999.3
|
WDR62
|
WD repeat domain 62 |
| chr17_+_30593195 | 1.05 |
ENST00000431505.2
ENST00000269051.4 ENST00000538145.1 |
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr16_-_54962415 | 1.03 |
ENST00000501177.3
ENST00000559598.2 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr17_-_4269920 | 1.02 |
ENST00000572484.1
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 |
| chrX_-_153602991 | 1.02 |
ENST00000369850.3
ENST00000422373.1 |
FLNA
|
filamin A, alpha |
| chr12_+_102271436 | 0.99 |
ENST00000544152.1
|
DRAM1
|
DNA-damage regulated autophagy modulator 1 |
| chr11_-_66336060 | 0.99 |
ENST00000310325.5
|
CTSF
|
cathepsin F |
| chr19_+_6372444 | 0.99 |
ENST00000245812.3
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli) |
| chr20_+_42295745 | 0.99 |
ENST00000396863.4
ENST00000217026.4 |
MYBL2
|
v-myb avian myeloblastosis viral oncogene homolog-like 2 |
| chr7_-_107643567 | 0.97 |
ENST00000393559.1
ENST00000439976.1 ENST00000393560.1 |
LAMB1
|
laminin, beta 1 |
| chr11_-_75062829 | 0.96 |
ENST00000393505.4
|
ARRB1
|
arrestin, beta 1 |
| chr10_+_94608245 | 0.94 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr16_-_30006922 | 0.93 |
ENST00000564026.1
|
HIRIP3
|
HIRA interacting protein 3 |
| chr1_+_861095 | 0.92 |
ENST00000342066.3
|
SAMD11
|
sterile alpha motif domain containing 11 |
| chr5_-_41510725 | 0.89 |
ENST00000328457.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr8_+_6565854 | 0.89 |
ENST00000285518.6
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 |
| chr12_+_58005204 | 0.88 |
ENST00000286494.4
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr12_+_12764773 | 0.86 |
ENST00000228865.2
|
CREBL2
|
cAMP responsive element binding protein-like 2 |
| chr16_-_54962625 | 0.85 |
ENST00000559432.1
|
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr1_+_212458834 | 0.85 |
ENST00000261461.2
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr2_+_30670077 | 0.84 |
ENST00000466477.1
ENST00000465200.1 ENST00000379509.3 ENST00000319406.4 ENST00000488144.1 ENST00000465538.1 ENST00000309052.4 ENST00000359433.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr10_-_118502070 | 0.84 |
ENST00000369209.3
|
HSPA12A
|
heat shock 70kDa protein 12A |
| chr9_+_100174232 | 0.84 |
ENST00000355295.4
|
TDRD7
|
tudor domain containing 7 |
| chr1_+_212208919 | 0.83 |
ENST00000366991.4
ENST00000542077.1 |
DTL
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr6_+_26104104 | 0.83 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr2_-_97523721 | 0.81 |
ENST00000393537.4
|
ANKRD39
|
ankyrin repeat domain 39 |
| chr5_-_138718973 | 0.78 |
ENST00000353963.3
ENST00000348729.3 |
SLC23A1
|
solute carrier family 23 (ascorbic acid transporter), member 1 |
| chr17_-_4269768 | 0.78 |
ENST00000396981.2
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr1_+_22351977 | 0.77 |
ENST00000420503.1
ENST00000416769.1 ENST00000404210.2 |
LINC00339
|
long intergenic non-protein coding RNA 339 |
| chr5_-_41510656 | 0.77 |
ENST00000377801.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr11_-_6440624 | 0.77 |
ENST00000311051.3
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr8_-_142318398 | 0.76 |
ENST00000520137.1
|
SLC45A4
|
solute carrier family 45, member 4 |
| chr6_+_24495067 | 0.74 |
ENST00000357578.3
ENST00000546278.1 ENST00000491546.1 |
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr16_-_87903079 | 0.74 |
ENST00000261622.4
|
SLC7A5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
| chrX_+_21392553 | 0.73 |
ENST00000279451.4
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr5_+_142149932 | 0.73 |
ENST00000274498.4
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr5_+_142149955 | 0.73 |
ENST00000378004.3
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr17_-_28619059 | 0.72 |
ENST00000580709.1
|
BLMH
|
bleomycin hydrolase |
| chr16_-_2162831 | 0.71 |
ENST00000483024.1
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant) |
| chr9_+_100174344 | 0.69 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr1_-_92351666 | 0.68 |
ENST00000465892.2
ENST00000417833.2 |
TGFBR3
|
transforming growth factor, beta receptor III |
| chr9_-_74383302 | 0.67 |
ENST00000377066.5
|
TMEM2
|
transmembrane protein 2 |
| chr8_-_11324273 | 0.66 |
ENST00000284486.4
|
FAM167A
|
family with sequence similarity 167, member A |
| chr6_+_159290917 | 0.66 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr14_+_75894714 | 0.65 |
ENST00000559060.1
|
JDP2
|
Jun dimerization protein 2 |
| chr2_+_28974489 | 0.65 |
ENST00000455580.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr11_+_1411129 | 0.64 |
ENST00000308219.9
ENST00000528841.1 ENST00000531197.1 ENST00000308230.5 |
BRSK2
|
BR serine/threonine kinase 2 |
| chr12_+_21654714 | 0.64 |
ENST00000542038.1
ENST00000540141.1 ENST00000229314.5 |
GOLT1B
|
golgi transport 1B |
| chr15_+_101459420 | 0.62 |
ENST00000388948.3
ENST00000284395.5 ENST00000534045.1 ENST00000532029.2 |
LRRK1
|
leucine-rich repeat kinase 1 |
| chr17_+_12693001 | 0.62 |
ENST00000262444.9
|
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr2_+_42396472 | 0.61 |
ENST00000318522.5
ENST00000402711.2 |
EML4
|
echinoderm microtubule associated protein like 4 |
| chr2_+_30670209 | 0.61 |
ENST00000497423.1
ENST00000476535.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr3_-_194207388 | 0.59 |
ENST00000457986.1
|
ATP13A3
|
ATPase type 13A3 |
| chr12_-_1703331 | 0.59 |
ENST00000339235.3
|
FBXL14
|
F-box and leucine-rich repeat protein 14 |
| chrX_-_119695279 | 0.57 |
ENST00000336592.6
|
CUL4B
|
cullin 4B |
| chr2_+_42396574 | 0.56 |
ENST00000401738.3
|
EML4
|
echinoderm microtubule associated protein like 4 |
| chr6_+_24495185 | 0.56 |
ENST00000348925.2
|
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr12_+_49761147 | 0.56 |
ENST00000549298.1
|
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr9_-_115095229 | 0.56 |
ENST00000210227.4
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr2_-_37384175 | 0.55 |
ENST00000411537.2
ENST00000233057.4 ENST00000395127.2 ENST00000390013.3 |
EIF2AK2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr11_+_66059339 | 0.55 |
ENST00000327259.4
|
TMEM151A
|
transmembrane protein 151A |
| chr17_-_1419878 | 0.54 |
ENST00000449479.1
ENST00000477910.1 ENST00000542125.1 ENST00000575172.1 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr16_+_84002234 | 0.53 |
ENST00000305202.4
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2 |
| chr17_-_42276574 | 0.52 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr1_-_94703118 | 0.52 |
ENST00000260526.6
ENST00000370217.3 |
ARHGAP29
|
Rho GTPase activating protein 29 |
| chr12_+_49760639 | 0.50 |
ENST00000549538.1
ENST00000548654.1 ENST00000550643.1 ENST00000548710.1 ENST00000549179.1 ENST00000548377.1 |
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr5_-_142783175 | 0.50 |
ENST00000231509.3
ENST00000394464.2 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr17_+_27895045 | 0.47 |
ENST00000580183.2
ENST00000578749.1 ENST00000582829.2 |
TP53I13
|
tumor protein p53 inducible protein 13 |
| chr2_-_175547571 | 0.46 |
ENST00000409415.3
ENST00000359761.3 ENST00000272746.5 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr5_+_176873446 | 0.46 |
ENST00000507881.1
|
PRR7
|
proline rich 7 (synaptic) |
| chr4_-_18023350 | 0.46 |
ENST00000539056.1
ENST00000382226.5 ENST00000326877.4 |
LCORL
|
ligand dependent nuclear receptor corepressor-like |
| chr1_-_92351769 | 0.46 |
ENST00000212355.4
|
TGFBR3
|
transforming growth factor, beta receptor III |
| chr22_+_29168652 | 0.45 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr10_+_94608218 | 0.45 |
ENST00000371543.1
|
EXOC6
|
exocyst complex component 6 |
| chr13_+_48877895 | 0.44 |
ENST00000267163.4
|
RB1
|
retinoblastoma 1 |
| chr15_-_35280426 | 0.44 |
ENST00000559564.1
ENST00000356321.4 |
ZNF770
|
zinc finger protein 770 |
| chr6_-_89673280 | 0.43 |
ENST00000369475.3
ENST00000369485.4 ENST00000538899.1 ENST00000265607.6 |
RNGTT
|
RNA guanylyltransferase and 5'-phosphatase |
| chr19_+_45596398 | 0.42 |
ENST00000544069.2
|
PPP1R37
|
protein phosphatase 1, regulatory subunit 37 |
| chrX_-_38186775 | 0.42 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr17_-_1419914 | 0.42 |
ENST00000397335.3
ENST00000574561.1 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr2_+_26915584 | 0.41 |
ENST00000302909.3
|
KCNK3
|
potassium channel, subfamily K, member 3 |
| chr13_+_48611665 | 0.41 |
ENST00000258662.2
|
NUDT15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr9_+_131085095 | 0.41 |
ENST00000372875.3
|
COQ4
|
coenzyme Q4 |
| chr11_+_1411503 | 0.41 |
ENST00000526678.1
|
BRSK2
|
BR serine/threonine kinase 2 |
| chr11_+_28131821 | 0.40 |
ENST00000379199.2
ENST00000303459.6 |
METTL15
|
methyltransferase like 15 |
| chr9_+_129622904 | 0.40 |
ENST00000319119.4
|
ZBTB34
|
zinc finger and BTB domain containing 34 |
| chr12_-_117175819 | 0.39 |
ENST00000261318.3
ENST00000536380.1 |
C12orf49
|
chromosome 12 open reading frame 49 |
| chr18_-_268019 | 0.39 |
ENST00000261600.6
|
THOC1
|
THO complex 1 |
| chr19_-_45004556 | 0.39 |
ENST00000587047.1
ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180
|
zinc finger protein 180 |
| chr15_-_75932528 | 0.38 |
ENST00000403490.1
|
IMP3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr16_-_2185899 | 0.38 |
ENST00000262304.4
ENST00000423118.1 |
PKD1
|
polycystic kidney disease 1 (autosomal dominant) |
| chr7_-_73668692 | 0.38 |
ENST00000352131.3
ENST00000055077.3 |
RFC2
|
replication factor C (activator 1) 2, 40kDa |
| chr6_-_111804905 | 0.37 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr3_+_133524459 | 0.36 |
ENST00000484684.1
|
SRPRB
|
signal recognition particle receptor, B subunit |
| chrX_-_38186811 | 0.36 |
ENST00000318842.7
|
RPGR
|
retinitis pigmentosa GTPase regulator |
| chrX_-_154299501 | 0.36 |
ENST00000369476.3
ENST00000369484.3 |
MTCP1
CMC4
|
mature T-cell proliferation 1 C-x(9)-C motif containing 4 |
| chr5_-_132166303 | 0.36 |
ENST00000440118.1
|
SHROOM1
|
shroom family member 1 |
| chr17_-_1420006 | 0.35 |
ENST00000320345.6
ENST00000406424.4 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr12_+_90102729 | 0.35 |
ENST00000605386.1
|
LINC00936
|
long intergenic non-protein coding RNA 936 |
| chr1_-_153919128 | 0.35 |
ENST00000361217.4
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr5_+_61602236 | 0.34 |
ENST00000514082.1
ENST00000407818.3 |
KIF2A
|
kinesin heavy chain member 2A |
| chr2_-_128785619 | 0.34 |
ENST00000450957.1
|
SAP130
|
Sin3A-associated protein, 130kDa |
| chr9_+_96214166 | 0.34 |
ENST00000375389.3
ENST00000333936.5 ENST00000340893.4 |
FAM120A
|
family with sequence similarity 120A |
| chrX_-_15353629 | 0.34 |
ENST00000333590.4
ENST00000428964.1 ENST00000542278.1 |
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A |
| chr16_+_30007524 | 0.33 |
ENST00000567254.1
ENST00000567705.1 |
INO80E
|
INO80 complex subunit E |
| chr19_-_49843539 | 0.33 |
ENST00000602554.1
ENST00000358234.4 |
CTC-301O7.4
|
CTC-301O7.4 |
| chr11_-_46867780 | 0.32 |
ENST00000529230.1
ENST00000415402.1 ENST00000312055.5 |
CKAP5
|
cytoskeleton associated protein 5 |
| chr22_+_23487513 | 0.32 |
ENST00000263116.2
ENST00000341989.4 |
RAB36
|
RAB36, member RAS oncogene family |
| chr17_-_1588101 | 0.32 |
ENST00000577001.1
ENST00000572621.1 ENST00000304992.6 |
PRPF8
|
pre-mRNA processing factor 8 |
| chr8_-_100905925 | 0.31 |
ENST00000518171.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr6_-_24721054 | 0.31 |
ENST00000378119.4
|
C6orf62
|
chromosome 6 open reading frame 62 |
| chr12_+_53773944 | 0.31 |
ENST00000551969.1
ENST00000327443.4 |
SP1
|
Sp1 transcription factor |
| chr19_+_45596218 | 0.31 |
ENST00000421905.1
ENST00000221462.4 |
PPP1R37
|
protein phosphatase 1, regulatory subunit 37 |
| chr17_-_1419941 | 0.30 |
ENST00000498390.1
|
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr14_-_101034811 | 0.30 |
ENST00000553553.1
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chrX_+_17393543 | 0.30 |
ENST00000380060.3
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
| chr9_-_115095123 | 0.30 |
ENST00000458258.1
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr9_-_98279241 | 0.29 |
ENST00000437951.1
ENST00000375274.2 ENST00000430669.2 ENST00000468211.2 |
PTCH1
|
patched 1 |
| chr4_+_108746282 | 0.29 |
ENST00000503862.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr11_+_3877412 | 0.29 |
ENST00000527651.1
|
STIM1
|
stromal interaction molecule 1 |
| chr14_+_75894391 | 0.28 |
ENST00000419727.2
|
JDP2
|
Jun dimerization protein 2 |
| chr2_+_64068116 | 0.28 |
ENST00000480679.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr13_-_99738867 | 0.28 |
ENST00000427887.1
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr17_-_46115122 | 0.28 |
ENST00000006101.4
|
COPZ2
|
coatomer protein complex, subunit zeta 2 |
| chr1_-_51796226 | 0.28 |
ENST00000451380.1
ENST00000371747.3 ENST00000439482.1 |
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr10_-_75532373 | 0.28 |
ENST00000595757.1
|
AC022400.2
|
Uncharacterized protein; cDNA FLJ44715 fis, clone BRACE3021430 |
| chr12_-_125052010 | 0.27 |
ENST00000458234.1
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr5_-_154317740 | 0.27 |
ENST00000285873.7
|
GEMIN5
|
gem (nuclear organelle) associated protein 5 |
| chr2_-_54087066 | 0.27 |
ENST00000394705.2
ENST00000352846.3 ENST00000406625.2 |
GPR75
GPR75-ASB3
ASB3
|
G protein-coupled receptor 75 GPR75-ASB3 readthrough Ankyrin repeat and SOCS box protein 3 |
| chr17_-_1420182 | 0.27 |
ENST00000421807.2
|
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr17_-_16395455 | 0.27 |
ENST00000409083.3
|
FAM211A
|
family with sequence similarity 211, member A |
| chr3_+_196295482 | 0.26 |
ENST00000440469.1
ENST00000311630.6 |
FBXO45
|
F-box protein 45 |
| chr5_+_61601965 | 0.26 |
ENST00000401507.3
|
KIF2A
|
kinesin heavy chain member 2A |
| chr1_-_200379180 | 0.26 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr16_+_30006997 | 0.26 |
ENST00000304516.7
|
INO80E
|
INO80 complex subunit E |
| chr17_-_7232585 | 0.26 |
ENST00000571887.1
ENST00000315614.7 ENST00000399464.2 ENST00000570460.1 |
NEURL4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr7_+_98972327 | 0.25 |
ENST00000455009.1
|
ARPC1B
|
actin related protein 2/3 complex, subunit 1B, 41kDa |
| chr3_-_28390120 | 0.25 |
ENST00000334100.6
|
AZI2
|
5-azacytidine induced 2 |
| chr13_+_25875785 | 0.25 |
ENST00000381747.3
|
NUPL1
|
nucleoporin like 1 |
| chr5_-_158526756 | 0.25 |
ENST00000313708.6
ENST00000517373.1 |
EBF1
|
early B-cell factor 1 |
| chr2_+_121010370 | 0.25 |
ENST00000420510.1
|
RALB
|
v-ral simian leukemia viral oncogene homolog B |
| chr2_+_120770581 | 0.25 |
ENST00000263713.5
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr14_+_64971438 | 0.25 |
ENST00000555321.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr11_+_64073699 | 0.25 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr11_-_73309112 | 0.25 |
ENST00000450446.2
|
FAM168A
|
family with sequence similarity 168, member A |
| chr10_-_62761188 | 0.24 |
ENST00000357917.4
|
RHOBTB1
|
Rho-related BTB domain containing 1 |
| chr17_-_77813186 | 0.24 |
ENST00000448310.1
ENST00000269397.4 |
CBX4
|
chromobox homolog 4 |
| chrX_-_153285251 | 0.24 |
ENST00000444230.1
ENST00000393682.1 ENST00000393687.2 ENST00000429936.2 ENST00000369974.2 |
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr2_+_204193149 | 0.24 |
ENST00000422511.2
|
ABI2
|
abl-interactor 2 |
| chr13_-_31039375 | 0.24 |
ENST00000399494.1
|
HMGB1
|
high mobility group box 1 |
| chr2_+_204193129 | 0.24 |
ENST00000417864.1
|
ABI2
|
abl-interactor 2 |
| chr3_-_13009168 | 0.23 |
ENST00000273221.4
|
IQSEC1
|
IQ motif and Sec7 domain 1 |
| chr13_-_48612067 | 0.23 |
ENST00000543413.1
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr9_-_72374848 | 0.22 |
ENST00000377200.5
ENST00000340434.4 ENST00000472967.2 |
PTAR1
|
protein prenyltransferase alpha subunit repeat containing 1 |
| chr5_+_61602055 | 0.22 |
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A |
| chr2_+_120770645 | 0.21 |
ENST00000443902.2
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chrX_-_25034065 | 0.21 |
ENST00000379044.4
|
ARX
|
aristaless related homeobox |
| chr4_-_77997126 | 0.21 |
ENST00000537948.1
ENST00000507788.1 ENST00000237654.4 |
CCNI
|
cyclin I |
| chr5_-_142782862 | 0.21 |
ENST00000415690.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr17_+_30771279 | 0.20 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr22_-_20255212 | 0.20 |
ENST00000416372.1
|
RTN4R
|
reticulon 4 receptor |
| chr16_-_70557430 | 0.20 |
ENST00000393612.4
ENST00000564653.1 ENST00000323786.5 |
COG4
|
component of oligomeric golgi complex 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HINFP
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 8.2 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.9 | 4.5 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.8 | 2.4 | GO:0042660 | positive regulation of cell fate specification(GO:0042660) |
| 0.7 | 2.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.6 | 1.9 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.5 | 1.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.4 | 1.3 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.4 | 2.2 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.4 | 1.4 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.4 | 3.2 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.3 | 1.0 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.3 | 1.9 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.3 | 1.1 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.3 | 2.7 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.3 | 0.8 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.3 | 1.3 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.2 | 3.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 1.2 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.2 | 1.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 | 1.7 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.2 | 1.0 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.2 | 0.8 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 | 1.0 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.2 | 0.6 | GO:0001928 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.2 | 1.7 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.1 | 0.4 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 | 0.4 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.1 | 0.5 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 1.2 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 0.1 | 0.9 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.6 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 1.3 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 0.3 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.7 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 1.5 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.3 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.1 | 0.4 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.1 | 0.4 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.1 | 0.2 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.1 | 1.5 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 | 0.2 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.1 | 0.4 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.3 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.3 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.2 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.1 | 0.6 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.4 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.7 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 1.0 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.0 | 2.4 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.9 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 1.0 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.4 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 1.5 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.3 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.7 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0006844 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.0 | 0.9 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.2 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.5 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.0 | 0.2 | GO:0070814 | phenotypic switching(GO:0036166) hydrogen sulfide biosynthetic process(GO:0070814) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.2 | GO:0021831 | embryonic olfactory bulb interneuron precursor migration(GO:0021831) |
| 0.0 | 0.6 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 1.8 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.7 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.4 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 1.9 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.0 | 1.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.3 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.4 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.6 | GO:0036035 | osteoclast development(GO:0036035) |
| 0.0 | 0.9 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 1.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.5 | 2.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.3 | 1.0 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.2 | 1.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 1.1 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.2 | 1.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 1.9 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.2 | 2.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 8.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.4 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 1.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 3.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.7 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 1.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 2.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.4 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 4.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 2.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.4 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 1.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.1 | GO:0005940 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.0 | 0.7 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 1.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.2 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.4 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.4 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.0 | 0.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.6 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.8 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.9 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.7 | 8.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.5 | 3.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.4 | 1.1 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.3 | 1.7 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.3 | 0.8 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.2 | 1.9 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 0.8 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 1.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 0.4 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.1 | 0.4 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.1 | 0.6 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.1 | 3.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 3.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 1.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.4 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 1.9 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 2.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.7 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 1.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.2 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.1 | 0.4 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.3 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 1.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.4 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 0.4 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.7 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 2.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.2 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.3 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.3 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.4 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.5 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 2.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.2 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.1 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.0 | 0.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 1.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 1.3 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 1.0 | GO:0060590 | ATPase regulator activity(GO:0060590) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 5.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 1.6 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 6.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 3.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 2.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.9 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 3.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.9 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 4.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 2.4 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 0.4 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 1.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 3.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 3.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.5 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 2.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 2.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.4 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 1.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 1.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |