Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
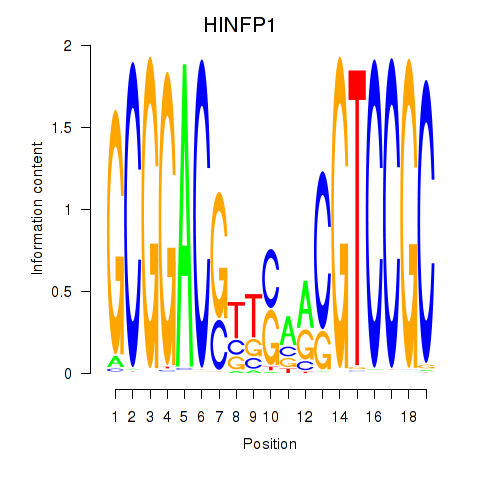
Results for HINFP1
Z-value: 0.25
Transcription factors associated with HINFP1
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity profile of HINFP1 motif
Sorted Z-values of HINFP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_2043689 | 0.81 |
ENST00000382878.3
ENST00000409248.4 |
C4orf48
|
chromosome 4 open reading frame 48 |
| chr2_-_234763105 | 0.62 |
ENST00000454020.1
|
HJURP
|
Holliday junction recognition protein |
| chr19_+_33685490 | 0.57 |
ENST00000253193.7
|
LRP3
|
low density lipoprotein receptor-related protein 3 |
| chr4_+_2043777 | 0.57 |
ENST00000409860.1
|
C4orf48
|
chromosome 4 open reading frame 48 |
| chr2_-_234763147 | 0.56 |
ENST00000411486.2
ENST00000432087.1 ENST00000441687.1 ENST00000414924.1 |
HJURP
|
Holliday junction recognition protein |
| chr2_+_130939235 | 0.55 |
ENST00000425361.1
ENST00000457492.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr2_+_130939827 | 0.49 |
ENST00000409255.1
ENST00000455239.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr10_+_72238517 | 0.46 |
ENST00000263563.6
|
PALD1
|
phosphatase domain containing, paladin 1 |
| chrX_-_153707246 | 0.39 |
ENST00000407062.1
|
LAGE3
|
L antigen family, member 3 |
| chr18_+_11981427 | 0.36 |
ENST00000269159.3
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr20_+_35974532 | 0.35 |
ENST00000373578.2
|
SRC
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr1_+_109102652 | 0.28 |
ENST00000370035.3
ENST00000405454.1 |
FAM102B
|
family with sequence similarity 102, member B |
| chr18_+_11981547 | 0.26 |
ENST00000588927.1
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr15_-_32162833 | 0.21 |
ENST00000560598.1
|
OTUD7A
|
OTU domain containing 7A |
| chr15_+_85525205 | 0.20 |
ENST00000394553.1
ENST00000339708.5 |
PDE8A
|
phosphodiesterase 8A |
| chr9_-_33264676 | 0.15 |
ENST00000472232.3
ENST00000379704.2 |
BAG1
|
BCL2-associated athanogene |
| chr7_+_1609694 | 0.14 |
ENST00000437621.2
ENST00000457484.2 |
PSMG3-AS1
|
PSMG3 antisense RNA 1 (head to head) |
| chr22_+_27053422 | 0.14 |
ENST00000413665.1
ENST00000421151.1 ENST00000456129.1 ENST00000430080.1 |
MIAT
|
myocardial infarction associated transcript (non-protein coding) |
| chr10_+_22614547 | 0.13 |
ENST00000416820.1
|
BMI1
|
BMI1 polycomb ring finger oncogene |
| chr6_-_144416737 | 0.13 |
ENST00000367569.2
|
SF3B5
|
splicing factor 3b, subunit 5, 10kDa |
| chr7_+_1609765 | 0.12 |
ENST00000437964.1
ENST00000533935.1 ENST00000532358.1 ENST00000524978.1 |
PSMG3-AS1
|
PSMG3 antisense RNA 1 (head to head) |
| chr17_-_79481666 | 0.11 |
ENST00000575659.1
|
ACTG1
|
actin, gamma 1 |
| chr7_-_1609591 | 0.09 |
ENST00000288607.2
ENST00000404674.3 |
PSMG3
|
proteasome (prosome, macropain) assembly chaperone 3 |
| chr6_-_168720382 | 0.08 |
ENST00000610183.1
ENST00000607983.1 ENST00000366795.3 |
DACT2
|
dishevelled-binding antagonist of beta-catenin 2 |
| chr2_-_130939115 | 0.07 |
ENST00000441135.1
ENST00000339679.7 ENST00000426662.2 ENST00000443958.2 ENST00000351288.6 ENST00000453750.1 ENST00000452225.2 |
SMPD4
|
sphingomyelin phosphodiesterase 4, neutral membrane (neutral sphingomyelinase-3) |
| chr10_-_97453650 | 0.07 |
ENST00000371209.5
ENST00000371217.5 ENST00000430368.2 |
TCTN3
|
tectonic family member 3 |
| chr3_-_196045127 | 0.07 |
ENST00000325318.5
|
TCTEX1D2
|
Tctex1 domain containing 2 |
| chr1_-_43232649 | 0.06 |
ENST00000372526.2
ENST00000236040.4 ENST00000296388.5 ENST00000397054.3 |
LEPRE1
|
leucine proline-enriched proteoglycan (leprecan) 1 |
| chr14_+_45366518 | 0.06 |
ENST00000557112.1
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr1_-_222886526 | 0.06 |
ENST00000541237.1
|
AIDA
|
axin interactor, dorsalization associated |
| chr11_+_637246 | 0.05 |
ENST00000176183.5
|
DRD4
|
dopamine receptor D4 |
| chr20_-_3996165 | 0.04 |
ENST00000545616.2
ENST00000358395.6 |
RNF24
|
ring finger protein 24 |
| chr10_-_977564 | 0.02 |
ENST00000406525.2
|
LARP4B
|
La ribonucleoprotein domain family, member 4B |
| chr1_-_42921915 | 0.02 |
ENST00000372565.3
ENST00000433602.2 |
ZMYND12
|
zinc finger, MYND-type containing 12 |
| chr6_-_52149475 | 0.02 |
ENST00000419835.2
ENST00000229854.7 ENST00000596288.1 |
MCM3
|
minichromosome maintenance complex component 3 |
| chr19_-_5680499 | 0.01 |
ENST00000587589.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr14_+_73704201 | 0.01 |
ENST00000340738.5
ENST00000427855.1 ENST00000381166.3 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
Network of associatons between targets according to the STRING database.
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.6 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 1.2 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.4 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST STAT3 PATHWAY | STAT3 Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |