Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
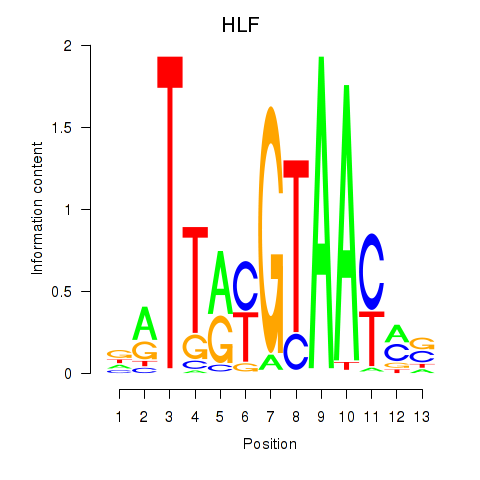
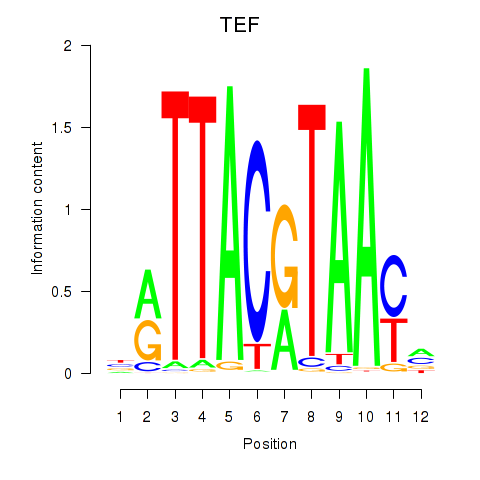
Results for HLF_TEF
Z-value: 0.71
Transcription factors associated with HLF_TEF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HLF
|
ENSG00000108924.9 | HLF transcription factor, PAR bZIP family member |
|
TEF
|
ENSG00000167074.10 | TEF transcription factor, PAR bZIP family member |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HLF | hg19_v2_chr17_+_53343171_53343207 | 0.65 | 1.8e-03 | Click! |
| TEF | hg19_v2_chr22_+_41777927_41777971 | -0.53 | 1.7e-02 | Click! |
Activity profile of HLF_TEF motif
Sorted Z-values of HLF_TEF motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_27469383 | 3.14 |
ENST00000519742.1
|
CLU
|
clusterin |
| chr8_-_27469196 | 2.80 |
ENST00000546343.1
ENST00000560566.1 |
CLU
|
clusterin |
| chr8_-_27468945 | 2.77 |
ENST00000405140.3
|
CLU
|
clusterin |
| chr8_-_27468842 | 2.76 |
ENST00000523500.1
|
CLU
|
clusterin |
| chr8_-_27468717 | 2.62 |
ENST00000520796.1
ENST00000520491.1 |
CLU
|
clusterin |
| chr2_-_240230890 | 1.58 |
ENST00000446876.1
|
HDAC4
|
histone deacetylase 4 |
| chr17_-_19651654 | 1.53 |
ENST00000395555.3
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr17_-_41984835 | 1.51 |
ENST00000520406.1
ENST00000518478.1 ENST00000522172.1 ENST00000461854.1 ENST00000521178.1 ENST00000520305.1 ENST00000523501.1 ENST00000520241.1 |
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr19_+_17413663 | 1.49 |
ENST00000594999.1
|
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr17_-_19651668 | 1.31 |
ENST00000494157.2
ENST00000225740.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr17_-_19651598 | 1.29 |
ENST00000570414.1
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr6_-_64029879 | 1.26 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr20_+_33759854 | 1.17 |
ENST00000216968.4
|
PROCR
|
protein C receptor, endothelial |
| chr12_+_52430894 | 1.15 |
ENST00000546842.1
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr9_-_34665983 | 1.14 |
ENST00000416454.1
ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5
|
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr3_+_101504200 | 1.11 |
ENST00000422132.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr1_-_226496772 | 1.10 |
ENST00000359525.2
ENST00000460719.1 |
LIN9
|
lin-9 homolog (C. elegans) |
| chr18_+_32621324 | 1.09 |
ENST00000300249.5
ENST00000538170.2 ENST00000588910.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr17_+_685513 | 0.99 |
ENST00000304478.4
|
RNMTL1
|
RNA methyltransferase like 1 |
| chr17_-_41623075 | 0.98 |
ENST00000545089.1
|
ETV4
|
ets variant 4 |
| chr14_-_23285069 | 0.92 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr1_-_244013384 | 0.92 |
ENST00000366539.1
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr5_-_58882219 | 0.87 |
ENST00000505453.1
ENST00000360047.5 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr17_-_41623691 | 0.82 |
ENST00000545954.1
|
ETV4
|
ets variant 4 |
| chr17_-_41623716 | 0.81 |
ENST00000319349.5
|
ETV4
|
ets variant 4 |
| chr17_-_41623259 | 0.78 |
ENST00000538265.1
ENST00000591713.1 |
ETV4
|
ets variant 4 |
| chr7_-_44887620 | 0.77 |
ENST00000349299.3
ENST00000521529.1 ENST00000308153.4 ENST00000350771.3 ENST00000222690.6 ENST00000381124.5 ENST00000437072.1 ENST00000446531.1 |
H2AFV
|
H2A histone family, member V |
| chr17_-_36956155 | 0.77 |
ENST00000269554.3
|
PIP4K2B
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta |
| chr14_-_23285011 | 0.74 |
ENST00000397532.3
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr3_-_158390282 | 0.73 |
ENST00000264265.3
|
LXN
|
latexin |
| chr11_+_69061594 | 0.73 |
ENST00000441339.2
ENST00000308946.3 ENST00000535407.1 |
MYEOV
|
myeloma overexpressed |
| chr21_+_45138941 | 0.73 |
ENST00000398081.1
ENST00000468090.1 ENST00000291565.4 |
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr5_+_172332220 | 0.73 |
ENST00000518247.1
ENST00000326654.2 |
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr16_-_54962704 | 0.72 |
ENST00000502066.2
ENST00000560912.1 ENST00000558952.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr7_+_23145884 | 0.71 |
ENST00000409689.1
ENST00000410047.1 |
KLHL7
|
kelch-like family member 7 |
| chr6_+_24126350 | 0.70 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr16_-_53537105 | 0.68 |
ENST00000568596.1
ENST00000570004.1 ENST00000564497.1 ENST00000300245.4 ENST00000394657.7 |
AKTIP
|
AKT interacting protein |
| chr17_-_26941179 | 0.68 |
ENST00000301037.5
ENST00000530121.1 ENST00000525510.1 ENST00000577790.1 ENST00000531839.1 ENST00000534850.1 |
SGK494
RP11-192H23.4
|
uncharacterized serine/threonine-protein kinase SgK494 Uncharacterized protein |
| chr9_-_123639445 | 0.66 |
ENST00000312189.6
|
PHF19
|
PHD finger protein 19 |
| chr1_+_154378049 | 0.66 |
ENST00000512471.1
|
IL6R
|
interleukin 6 receptor |
| chr9_-_123639304 | 0.65 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19 |
| chr1_+_45212051 | 0.64 |
ENST00000372222.3
|
KIF2C
|
kinesin family member 2C |
| chr8_+_90914073 | 0.63 |
ENST00000297438.2
|
OSGIN2
|
oxidative stress induced growth inhibitor family member 2 |
| chr14_-_23284703 | 0.61 |
ENST00000555911.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr12_+_52431016 | 0.61 |
ENST00000553200.1
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr2_-_225362533 | 0.60 |
ENST00000451538.1
|
CUL3
|
cullin 3 |
| chr1_+_45212074 | 0.60 |
ENST00000372217.1
|
KIF2C
|
kinesin family member 2C |
| chr2_-_167232484 | 0.59 |
ENST00000375387.4
ENST00000303354.6 ENST00000409672.1 |
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit |
| chr16_-_54962415 | 0.56 |
ENST00000501177.3
ENST00000559598.2 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr9_-_130712995 | 0.55 |
ENST00000373084.4
|
FAM102A
|
family with sequence similarity 102, member A |
| chr17_+_73089382 | 0.54 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr10_+_90660832 | 0.54 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr8_+_110552337 | 0.53 |
ENST00000337573.5
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr17_-_41623009 | 0.53 |
ENST00000393664.2
|
ETV4
|
ets variant 4 |
| chr3_+_141103634 | 0.51 |
ENST00000507722.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr17_-_685559 | 0.50 |
ENST00000301329.6
|
GLOD4
|
glyoxalase domain containing 4 |
| chr5_-_140013275 | 0.50 |
ENST00000512545.1
ENST00000302014.6 ENST00000401743.2 |
CD14
|
CD14 molecule |
| chr1_+_90308981 | 0.50 |
ENST00000527156.1
|
LRRC8D
|
leucine rich repeat containing 8 family, member D |
| chr1_+_169079823 | 0.49 |
ENST00000367813.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr2_+_183989157 | 0.49 |
ENST00000541912.1
|
NUP35
|
nucleoporin 35kDa |
| chr2_+_183989083 | 0.49 |
ENST00000295119.4
|
NUP35
|
nucleoporin 35kDa |
| chr8_+_19536083 | 0.49 |
ENST00000519803.1
|
RP11-1105O14.1
|
RP11-1105O14.1 |
| chr9_-_123639600 | 0.47 |
ENST00000373896.3
|
PHF19
|
PHD finger protein 19 |
| chr17_-_73266616 | 0.46 |
ENST00000579194.1
ENST00000581777.1 |
MIF4GD
|
MIF4G domain containing |
| chr4_+_75230853 | 0.45 |
ENST00000244869.2
|
EREG
|
epiregulin |
| chr14_+_102276132 | 0.44 |
ENST00000350249.3
ENST00000557621.1 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr3_-_45883558 | 0.43 |
ENST00000445698.1
ENST00000296135.6 |
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr9_-_88356789 | 0.43 |
ENST00000357081.3
ENST00000376081.4 ENST00000337006.4 ENST00000376109.3 |
AGTPBP1
|
ATP/GTP binding protein 1 |
| chr17_-_685493 | 0.42 |
ENST00000536578.1
ENST00000301328.5 ENST00000576419.1 |
GLOD4
|
glyoxalase domain containing 4 |
| chr7_-_15601595 | 0.42 |
ENST00000342526.3
|
AGMO
|
alkylglycerol monooxygenase |
| chrX_+_9431324 | 0.42 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr20_-_35329063 | 0.41 |
ENST00000422536.1
|
NDRG3
|
NDRG family member 3 |
| chr15_+_49913201 | 0.41 |
ENST00000329873.5
ENST00000558653.1 ENST00000559164.1 ENST00000560632.1 ENST00000559405.1 ENST00000251250.6 |
DTWD1
|
DTW domain containing 1 |
| chr17_+_73455788 | 0.41 |
ENST00000581519.1
|
KIAA0195
|
KIAA0195 |
| chr15_+_63050785 | 0.40 |
ENST00000472902.1
|
TLN2
|
talin 2 |
| chr12_-_122017542 | 0.38 |
ENST00000446152.2
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr20_-_33732952 | 0.38 |
ENST00000541621.1
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr13_-_45010939 | 0.38 |
ENST00000261489.2
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr8_+_94752349 | 0.37 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr8_-_102216925 | 0.36 |
ENST00000517844.1
|
ZNF706
|
zinc finger protein 706 |
| chr2_-_46769694 | 0.33 |
ENST00000522587.1
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr2_-_112614424 | 0.32 |
ENST00000427997.1
|
ANAPC1
|
anaphase promoting complex subunit 1 |
| chr11_+_105948216 | 0.32 |
ENST00000278618.4
|
AASDHPPT
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr16_+_70332956 | 0.32 |
ENST00000288071.6
ENST00000393657.2 ENST00000355992.3 ENST00000567706.1 |
DDX19B
RP11-529K1.3
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 19B Uncharacterized protein |
| chr1_-_226496898 | 0.30 |
ENST00000481685.1
|
LIN9
|
lin-9 homolog (C. elegans) |
| chr14_+_96722539 | 0.30 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr5_+_134303591 | 0.30 |
ENST00000282611.6
|
CATSPER3
|
cation channel, sperm associated 3 |
| chr16_-_70323422 | 0.30 |
ENST00000261772.8
|
AARS
|
alanyl-tRNA synthetase |
| chr19_+_1450112 | 0.29 |
ENST00000590469.1
ENST00000233607.2 ENST00000238483.4 ENST00000590877.1 |
APC2
|
adenomatosis polyposis coli 2 |
| chr14_-_23284675 | 0.29 |
ENST00000555959.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr5_-_43557791 | 0.29 |
ENST00000338972.4
ENST00000511321.1 ENST00000515338.1 |
PAIP1
|
poly(A) binding protein interacting protein 1 |
| chr3_+_62304648 | 0.28 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chr8_+_42396936 | 0.28 |
ENST00000416469.2
|
SMIM19
|
small integral membrane protein 19 |
| chr2_+_46769798 | 0.28 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr9_-_88356733 | 0.27 |
ENST00000376083.3
|
AGTPBP1
|
ATP/GTP binding protein 1 |
| chr12_+_48592134 | 0.27 |
ENST00000595310.1
|
DKFZP779L1853
|
DKFZP779L1853 |
| chr20_-_56285595 | 0.26 |
ENST00000395816.3
ENST00000347215.4 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr12_+_93963590 | 0.26 |
ENST00000340600.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr2_+_65663812 | 0.26 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr12_-_120966943 | 0.26 |
ENST00000552443.1
ENST00000547736.1 ENST00000445328.2 ENST00000547943.1 ENST00000288532.6 |
COQ5
|
coenzyme Q5 homolog, methyltransferase (S. cerevisiae) |
| chr17_+_70036164 | 0.25 |
ENST00000602013.1
|
AC007461.1
|
Uncharacterized protein |
| chr22_+_48972118 | 0.25 |
ENST00000358295.5
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5 |
| chr14_+_32476072 | 0.24 |
ENST00000556949.1
|
RP11-187E13.2
|
Uncharacterized protein |
| chr19_+_33865218 | 0.24 |
ENST00000585933.2
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr13_-_28545276 | 0.21 |
ENST00000381020.7
|
CDX2
|
caudal type homeobox 2 |
| chr3_-_58200398 | 0.21 |
ENST00000318316.3
ENST00000460422.1 ENST00000483681.1 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr3_+_158787041 | 0.21 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr5_-_140013224 | 0.21 |
ENST00000498971.2
|
CD14
|
CD14 molecule |
| chr4_+_157997273 | 0.21 |
ENST00000541722.1
ENST00000512619.1 |
GLRB
|
glycine receptor, beta |
| chr3_+_186330712 | 0.20 |
ENST00000411641.2
ENST00000273784.5 |
AHSG
|
alpha-2-HS-glycoprotein |
| chr14_+_75894714 | 0.20 |
ENST00000559060.1
|
JDP2
|
Jun dimerization protein 2 |
| chr16_-_54962625 | 0.19 |
ENST00000559432.1
|
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr19_-_46389359 | 0.19 |
ENST00000302165.3
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1 |
| chr7_+_23146271 | 0.19 |
ENST00000545771.1
|
KLHL7
|
kelch-like family member 7 |
| chr15_+_81293254 | 0.19 |
ENST00000267984.2
|
MESDC1
|
mesoderm development candidate 1 |
| chr4_+_96012614 | 0.19 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr4_+_48807155 | 0.19 |
ENST00000504654.1
|
OCIAD1
|
OCIA domain containing 1 |
| chr3_+_62304712 | 0.19 |
ENST00000494481.1
|
C3orf14
|
chromosome 3 open reading frame 14 |
| chr14_+_65453432 | 0.19 |
ENST00000246166.2
|
FNTB
|
farnesyltransferase, CAAX box, beta |
| chr1_-_114429997 | 0.19 |
ENST00000471267.1
ENST00000393320.3 |
BCL2L15
|
BCL2-like 15 |
| chr3_+_148447887 | 0.19 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr7_-_37026108 | 0.18 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr1_-_114430169 | 0.18 |
ENST00000393316.3
|
BCL2L15
|
BCL2-like 15 |
| chr2_+_102608306 | 0.18 |
ENST00000332549.3
|
IL1R2
|
interleukin 1 receptor, type II |
| chr17_-_41322332 | 0.18 |
ENST00000590740.1
|
RP11-242D8.1
|
RP11-242D8.1 |
| chr17_+_76037081 | 0.18 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr12_-_122884553 | 0.17 |
ENST00000535290.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr16_+_70680439 | 0.17 |
ENST00000288098.2
|
IL34
|
interleukin 34 |
| chr9_-_128246769 | 0.17 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr14_+_75894391 | 0.17 |
ENST00000419727.2
|
JDP2
|
Jun dimerization protein 2 |
| chr9_+_110045537 | 0.17 |
ENST00000358015.3
|
RAD23B
|
RAD23 homolog B (S. cerevisiae) |
| chr5_-_54830871 | 0.17 |
ENST00000307259.8
|
PPAP2A
|
phosphatidic acid phosphatase type 2A |
| chr7_-_45026200 | 0.17 |
ENST00000577700.1
ENST00000580458.1 ENST00000579383.1 ENST00000584686.1 ENST00000585030.1 ENST00000582727.1 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr19_-_41945804 | 0.16 |
ENST00000221943.9
ENST00000597457.1 ENST00000589970.1 ENST00000595425.1 ENST00000438807.3 ENST00000589102.1 ENST00000592922.2 |
ATP5SL
|
ATP5S-like |
| chr19_+_52076425 | 0.16 |
ENST00000436511.2
|
ZNF175
|
zinc finger protein 175 |
| chr10_-_82049424 | 0.16 |
ENST00000372213.3
|
MAT1A
|
methionine adenosyltransferase I, alpha |
| chr14_+_60716159 | 0.16 |
ENST00000325658.3
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr7_+_20686946 | 0.16 |
ENST00000443026.2
ENST00000406935.1 |
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr2_+_103378472 | 0.15 |
ENST00000412401.2
|
TMEM182
|
transmembrane protein 182 |
| chr4_+_41937131 | 0.15 |
ENST00000504986.1
ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33
|
transmembrane protein 33 |
| chr1_+_229440129 | 0.14 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr17_-_61850894 | 0.14 |
ENST00000403162.3
ENST00000582252.1 ENST00000225726.5 |
CCDC47
|
coiled-coil domain containing 47 |
| chr1_+_65730385 | 0.14 |
ENST00000263441.7
ENST00000395325.3 |
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr4_+_139694701 | 0.14 |
ENST00000502606.1
|
RP11-98O2.1
|
RP11-98O2.1 |
| chr2_-_99279928 | 0.13 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr5_+_12574944 | 0.13 |
ENST00000505877.1
ENST00000513051.1 ENST00000505196.1 |
CT49
|
cancer/testis antigen 49 (non-protein coding) |
| chr3_+_157154578 | 0.13 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr18_+_77160282 | 0.13 |
ENST00000318065.5
ENST00000545796.1 ENST00000592223.1 ENST00000329101.4 ENST00000586434.1 |
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr14_-_22005343 | 0.13 |
ENST00000327430.3
|
SALL2
|
spalt-like transcription factor 2 |
| chr3_+_188817891 | 0.13 |
ENST00000412373.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr19_-_1155118 | 0.13 |
ENST00000590998.1
|
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr7_-_45026159 | 0.13 |
ENST00000584327.1
ENST00000438705.3 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr7_+_135347215 | 0.13 |
ENST00000507606.1
|
C7orf73
|
chromosome 7 open reading frame 73 |
| chr12_-_53074182 | 0.12 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr21_+_35107346 | 0.12 |
ENST00000456489.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr21_+_35736302 | 0.12 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr19_+_50922187 | 0.12 |
ENST00000595883.1
ENST00000597855.1 ENST00000596074.1 ENST00000439922.2 ENST00000594685.1 ENST00000270632.7 |
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr10_-_65028817 | 0.11 |
ENST00000542921.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr9_+_27524283 | 0.11 |
ENST00000276943.2
|
IFNK
|
interferon, kappa |
| chr3_+_88188254 | 0.11 |
ENST00000309495.5
|
ZNF654
|
zinc finger protein 654 |
| chr10_-_65028938 | 0.11 |
ENST00000402544.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr12_-_12714006 | 0.10 |
ENST00000541207.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr5_+_95998070 | 0.10 |
ENST00000421689.2
ENST00000510756.1 ENST00000512620.1 |
CAST
|
calpastatin |
| chr9_+_101705893 | 0.10 |
ENST00000375001.3
|
COL15A1
|
collagen, type XV, alpha 1 |
| chr3_-_190580404 | 0.10 |
ENST00000442080.1
|
GMNC
|
geminin coiled-coil domain containing |
| chr9_+_136399929 | 0.10 |
ENST00000393060.1
|
ADAMTSL2
|
ADAMTS-like 2 |
| chr10_+_94050913 | 0.10 |
ENST00000358935.2
|
MARCH5
|
membrane-associated ring finger (C3HC4) 5 |
| chr11_-_105948129 | 0.10 |
ENST00000526793.1
|
KBTBD3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chr14_+_60715928 | 0.09 |
ENST00000395076.4
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr11_+_62495997 | 0.09 |
ENST00000316461.4
|
TTC9C
|
tetratricopeptide repeat domain 9C |
| chr6_-_46459099 | 0.08 |
ENST00000371374.1
|
RCAN2
|
regulator of calcineurin 2 |
| chr7_-_45026419 | 0.08 |
ENST00000578968.1
ENST00000580528.1 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr5_-_140013241 | 0.08 |
ENST00000519715.1
|
CD14
|
CD14 molecule |
| chr11_-_105948040 | 0.07 |
ENST00000534815.1
|
KBTBD3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chr12_+_57623477 | 0.07 |
ENST00000557487.1
ENST00000555634.1 ENST00000556689.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr14_-_72458326 | 0.07 |
ENST00000542853.1
|
AC005477.1
|
AC005477.1 |
| chr4_+_96012585 | 0.07 |
ENST00000502683.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr1_+_75594119 | 0.07 |
ENST00000294638.5
|
LHX8
|
LIM homeobox 8 |
| chr6_-_116447283 | 0.07 |
ENST00000452729.1
ENST00000243222.4 |
COL10A1
|
collagen, type X, alpha 1 |
| chr6_-_86353510 | 0.07 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr4_-_96470350 | 0.07 |
ENST00000504962.1
ENST00000453304.1 ENST00000506749.1 |
UNC5C
|
unc-5 homolog C (C. elegans) |
| chr7_-_73153178 | 0.07 |
ENST00000437775.2
ENST00000222800.3 |
ABHD11
|
abhydrolase domain containing 11 |
| chr6_+_97010424 | 0.06 |
ENST00000541107.1
ENST00000450218.1 ENST00000326771.2 |
FHL5
|
four and a half LIM domains 5 |
| chr12_+_57624085 | 0.06 |
ENST00000553474.1
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr7_-_141401951 | 0.06 |
ENST00000536163.1
|
KIAA1147
|
KIAA1147 |
| chr12_-_12714025 | 0.06 |
ENST00000539940.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr16_+_3508096 | 0.06 |
ENST00000577013.1
ENST00000570819.1 |
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr12_-_75603643 | 0.06 |
ENST00000549446.1
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr13_-_29292956 | 0.06 |
ENST00000266943.6
|
SLC46A3
|
solute carrier family 46, member 3 |
| chr17_+_61851157 | 0.05 |
ENST00000578681.1
ENST00000583590.1 |
DDX42
|
DEAD (Asp-Glu-Ala-Asp) box helicase 42 |
| chr17_-_34524157 | 0.05 |
ENST00000378354.4
ENST00000394484.1 |
CCL3L3
|
chemokine (C-C motif) ligand 3-like 3 |
| chr21_+_19617140 | 0.05 |
ENST00000299295.2
ENST00000338326.3 |
CHODL
|
chondrolectin |
| chr15_+_58430368 | 0.05 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr15_+_58430567 | 0.05 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr13_+_28813645 | 0.05 |
ENST00000282391.5
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr6_+_72926145 | 0.05 |
ENST00000425662.2
ENST00000453976.2 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr12_-_88423164 | 0.05 |
ENST00000298699.2
ENST00000550553.1 |
C12orf50
|
chromosome 12 open reading frame 50 |
| chr5_+_95997769 | 0.05 |
ENST00000338252.3
ENST00000508830.1 |
CAST
|
calpastatin |
| chr6_-_74231303 | 0.05 |
ENST00000309268.6
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr8_+_110656344 | 0.05 |
ENST00000499579.1
|
RP11-422N16.3
|
Uncharacterized protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of HLF_TEF
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 14.1 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.3 | 1.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.3 | 2.6 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.2 | 0.7 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.1 | 0.4 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 1.8 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 1.8 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.6 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.8 | GO:0071727 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 1.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.7 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.7 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 0.9 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.7 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.3 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 1.6 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.4 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.1 | 0.4 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.2 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.8 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.9 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.2 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.2 | GO:0045209 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.2 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 4.1 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.3 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.7 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.9 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.3 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.4 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:0070884 | calcineurin-NFAT signaling cascade(GO:0033173) regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.5 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.4 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 1.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.7 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.2 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 14.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 0.8 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.7 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.2 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.1 | 0.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 1.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.0 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 2.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 3.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.5 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.4 | 14.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.3 | 1.3 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 0.7 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.2 | 0.8 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 1.6 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 0.6 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 2.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.3 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.1 | 1.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 0.8 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.3 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.9 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 1.1 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.2 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.0 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.5 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 1.8 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 9.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 3.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 13.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.9 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 3.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.8 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 1.9 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 1.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.0 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |