Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
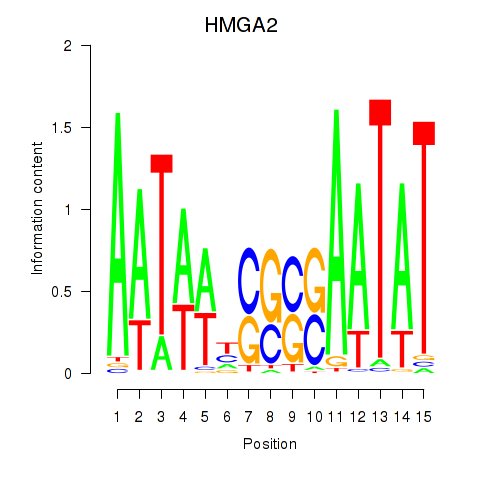
Results for HMGA2
Z-value: 0.56
Transcription factors associated with HMGA2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMGA2
|
ENSG00000149948.9 | high mobility group AT-hook 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMGA2 | hg19_v2_chr12_+_66218903_66218913 | -0.64 | 2.2e-03 | Click! |
Activity profile of HMGA2 motif
Sorted Z-values of HMGA2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_193853927 | 1.57 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr1_+_196621002 | 1.46 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr1_+_196621156 | 0.93 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr19_-_36297632 | 0.92 |
ENST00000588266.2
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr19_+_11457175 | 0.86 |
ENST00000458408.1
ENST00000586451.1 ENST00000588592.1 |
CCDC159
|
coiled-coil domain containing 159 |
| chr15_+_43985084 | 0.75 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr2_-_216257849 | 0.73 |
ENST00000456923.1
|
FN1
|
fibronectin 1 |
| chr15_+_43885252 | 0.73 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chrX_+_2670066 | 0.72 |
ENST00000381174.5
ENST00000419513.2 ENST00000426774.1 |
XG
|
Xg blood group |
| chr1_+_104104379 | 0.67 |
ENST00000435302.1
|
AMY2B
|
amylase, alpha 2B (pancreatic) |
| chr15_+_84904525 | 0.60 |
ENST00000510439.2
|
GOLGA6L4
|
golgin A6 family-like 4 |
| chr1_+_2487800 | 0.47 |
ENST00000355716.4
|
TNFRSF14
|
tumor necrosis factor receptor superfamily, member 14 |
| chrX_-_7895755 | 0.44 |
ENST00000444736.1
ENST00000537427.1 ENST00000442940.1 |
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chrX_+_135614293 | 0.42 |
ENST00000370634.3
|
VGLL1
|
vestigial like 1 (Drosophila) |
| chr12_-_10573149 | 0.42 |
ENST00000381904.2
ENST00000381903.2 ENST00000396439.2 |
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr3_+_121774202 | 0.41 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr19_+_41882466 | 0.38 |
ENST00000436170.2
|
TMEM91
|
transmembrane protein 91 |
| chrX_-_7895479 | 0.36 |
ENST00000381042.4
|
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr19_+_44455368 | 0.36 |
ENST00000591168.1
ENST00000587682.1 ENST00000251269.5 |
ZNF221
|
zinc finger protein 221 |
| chrX_+_1710484 | 0.33 |
ENST00000313871.3
ENST00000381261.3 |
AKAP17A
|
A kinase (PRKA) anchor protein 17A |
| chr15_+_83098710 | 0.32 |
ENST00000561062.1
ENST00000358583.3 |
GOLGA6L9
|
golgin A6 family-like 20 |
| chrX_-_71792477 | 0.31 |
ENST00000421523.1
ENST00000415409.1 ENST00000373559.4 ENST00000373556.3 ENST00000373560.2 ENST00000373583.1 ENST00000429103.2 ENST00000373571.1 ENST00000373554.1 |
HDAC8
|
histone deacetylase 8 |
| chr6_+_88106840 | 0.30 |
ENST00000369570.4
|
C6orf164
|
chromosome 6 open reading frame 164 |
| chr17_-_26220366 | 0.30 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr9_+_42704004 | 0.28 |
ENST00000457288.1
|
CBWD7
|
COBW domain containing 7 |
| chr15_-_82939157 | 0.28 |
ENST00000559949.1
|
RP13-996F3.5
|
golgin A6 family-like 18 |
| chr11_+_73661364 | 0.28 |
ENST00000339764.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr15_+_82722225 | 0.27 |
ENST00000300515.8
|
GOLGA6L9
|
golgin A6 family-like 9 |
| chr6_-_32908792 | 0.27 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr6_+_111580508 | 0.25 |
ENST00000368847.4
|
KIAA1919
|
KIAA1919 |
| chr10_-_62332357 | 0.25 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr2_+_220143989 | 0.25 |
ENST00000336576.5
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr1_+_144989309 | 0.25 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chrX_+_2670153 | 0.24 |
ENST00000509484.2
|
XG
|
Xg blood group |
| chr20_+_48884002 | 0.24 |
ENST00000425497.1
ENST00000445003.1 |
RP11-290F20.3
|
RP11-290F20.3 |
| chr12_-_10541575 | 0.23 |
ENST00000540818.1
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr19_-_11456722 | 0.23 |
ENST00000354882.5
|
TMEM205
|
transmembrane protein 205 |
| chr19_-_12267524 | 0.23 |
ENST00000455799.1
ENST00000355738.1 ENST00000439556.2 ENST00000542938.1 |
ZNF625
|
zinc finger protein 625 |
| chr6_+_116892641 | 0.22 |
ENST00000487832.2
ENST00000518117.1 |
RWDD1
|
RWD domain containing 1 |
| chr3_-_106959424 | 0.22 |
ENST00000607801.1
ENST00000479612.2 ENST00000484698.1 ENST00000477210.2 ENST00000473636.1 |
LINC00882
|
long intergenic non-protein coding RNA 882 |
| chr4_-_299110 | 0.22 |
ENST00000419098.1
|
ZNF732
|
zinc finger protein 732 |
| chr15_-_83018198 | 0.22 |
ENST00000557886.1
|
RP13-996F3.4
|
golgin A6 family-like 19 |
| chr19_-_11457082 | 0.22 |
ENST00000587948.1
|
TMEM205
|
transmembrane protein 205 |
| chrX_-_134186144 | 0.21 |
ENST00000370775.2
|
FAM127B
|
family with sequence similarity 127, member B |
| chr1_+_2487631 | 0.20 |
ENST00000409119.1
|
TNFRSF14
|
tumor necrosis factor receptor superfamily, member 14 |
| chr18_+_11851383 | 0.20 |
ENST00000526991.2
|
CHMP1B
|
charged multivesicular body protein 1B |
| chr19_-_11457162 | 0.20 |
ENST00000590482.1
|
TMEM205
|
transmembrane protein 205 |
| chr19_-_11456935 | 0.19 |
ENST00000590788.1
ENST00000586590.1 ENST00000589555.1 ENST00000586956.1 ENST00000593256.2 ENST00000447337.1 ENST00000591677.1 ENST00000586701.1 ENST00000589655.1 |
TMEM205
RAB3D
|
transmembrane protein 205 RAB3D, member RAS oncogene family |
| chr8_-_105479270 | 0.19 |
ENST00000521573.2
ENST00000351513.2 |
DPYS
|
dihydropyrimidinase |
| chr11_-_130786400 | 0.18 |
ENST00000265909.4
|
SNX19
|
sorting nexin 19 |
| chr2_-_239140011 | 0.18 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chr14_+_99947715 | 0.17 |
ENST00000389879.5
ENST00000557441.1 ENST00000555049.1 ENST00000555842.1 |
CCNK
|
cyclin K |
| chr14_+_50291993 | 0.17 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chrX_+_2924654 | 0.17 |
ENST00000381130.2
|
ARSH
|
arylsulfatase family, member H |
| chr12_+_65996599 | 0.16 |
ENST00000539116.1
ENST00000541391.1 |
RP11-221N13.3
|
RP11-221N13.3 |
| chr11_+_67250490 | 0.16 |
ENST00000528641.2
ENST00000279146.3 |
AIP
|
aryl hydrocarbon receptor interacting protein |
| chr5_+_36608422 | 0.16 |
ENST00000381918.3
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr6_-_32908765 | 0.16 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr3_+_186501336 | 0.16 |
ENST00000323963.5
ENST00000440191.2 ENST00000356531.5 |
EIF4A2
|
eukaryotic translation initiation factor 4A2 |
| chr14_-_65409438 | 0.16 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr2_+_177134134 | 0.16 |
ENST00000249442.6
ENST00000392529.2 ENST00000443241.1 |
MTX2
|
metaxin 2 |
| chr6_+_123110302 | 0.16 |
ENST00000368440.4
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr2_-_114461655 | 0.16 |
ENST00000424612.1
|
AC017074.2
|
AC017074.2 |
| chrY_+_22918021 | 0.16 |
ENST00000288666.5
|
RPS4Y2
|
ribosomal protein S4, Y-linked 2 |
| chr2_-_287299 | 0.15 |
ENST00000405290.1
|
FAM150B
|
family with sequence similarity 150, member B |
| chr2_+_13677795 | 0.15 |
ENST00000434509.1
|
AC092635.1
|
AC092635.1 |
| chr12_+_96196875 | 0.15 |
ENST00000553095.1
|
RP11-536G4.1
|
Uncharacterized protein |
| chr14_+_37641012 | 0.15 |
ENST00000556667.1
|
SLC25A21-AS1
|
SLC25A21 antisense RNA 1 |
| chr11_-_18063973 | 0.15 |
ENST00000528338.1
|
TPH1
|
tryptophan hydroxylase 1 |
| chr21_-_31852663 | 0.14 |
ENST00000390689.2
|
KRTAP19-1
|
keratin associated protein 19-1 |
| chr14_-_94759408 | 0.14 |
ENST00000554723.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr15_+_58702742 | 0.14 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr1_-_67600639 | 0.14 |
ENST00000544837.1
ENST00000603691.1 |
C1orf141
|
chromosome 1 open reading frame 141 |
| chr8_+_9911778 | 0.14 |
ENST00000317173.4
ENST00000441698.2 |
MSRA
|
methionine sulfoxide reductase A |
| chr14_-_94759595 | 0.13 |
ENST00000261994.4
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr14_+_88852059 | 0.13 |
ENST00000045347.7
|
SPATA7
|
spermatogenesis associated 7 |
| chr10_+_90484301 | 0.13 |
ENST00000404190.1
|
LIPK
|
lipase, family member K |
| chr5_-_135290705 | 0.12 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr5_+_67485704 | 0.12 |
ENST00000520762.1
|
RP11-404L6.2
|
Uncharacterized protein |
| chr14_-_94759361 | 0.12 |
ENST00000393096.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr15_-_32747835 | 0.12 |
ENST00000509311.2
ENST00000414865.2 |
GOLGA8O
|
golgin A8 family, member O |
| chr5_-_135290651 | 0.12 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr1_+_2487402 | 0.11 |
ENST00000451778.1
|
TNFRSF14
|
tumor necrosis factor receptor superfamily, member 14 |
| chr14_+_45553296 | 0.11 |
ENST00000355765.6
ENST00000553605.1 |
PRPF39
|
pre-mRNA processing factor 39 |
| chr2_-_183387430 | 0.11 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr2_-_183387283 | 0.11 |
ENST00000435564.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr20_+_9966728 | 0.10 |
ENST00000449270.1
|
RP5-839B4.8
|
RP5-839B4.8 |
| chr12_+_18414446 | 0.10 |
ENST00000433979.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
| chr19_-_11456872 | 0.09 |
ENST00000586218.1
|
TMEM205
|
transmembrane protein 205 |
| chr19_-_11456905 | 0.09 |
ENST00000588560.1
ENST00000592952.1 |
TMEM205
|
transmembrane protein 205 |
| chr7_+_80267973 | 0.09 |
ENST00000394788.3
ENST00000447544.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr10_-_129691195 | 0.09 |
ENST00000368671.3
|
CLRN3
|
clarin 3 |
| chr2_-_183387064 | 0.08 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr12_-_21757774 | 0.08 |
ENST00000261195.2
|
GYS2
|
glycogen synthase 2 (liver) |
| chrX_-_153523462 | 0.07 |
ENST00000361930.3
ENST00000369926.1 |
TEX28
|
testis expressed 28 |
| chr2_+_220144052 | 0.07 |
ENST00000425450.1
ENST00000392086.4 ENST00000421532.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr7_+_80275953 | 0.07 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr12_-_8765446 | 0.07 |
ENST00000537228.1
ENST00000229335.6 |
AICDA
|
activation-induced cytidine deaminase |
| chr9_-_86955598 | 0.07 |
ENST00000376238.4
|
SLC28A3
|
solute carrier family 28 (concentrative nucleoside transporter), member 3 |
| chr9_-_34397800 | 0.06 |
ENST00000297623.2
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr12_-_10324716 | 0.06 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr7_-_102985288 | 0.06 |
ENST00000379263.3
|
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chrX_-_128782722 | 0.05 |
ENST00000427399.1
|
APLN
|
apelin |
| chr1_+_55464600 | 0.05 |
ENST00000371265.4
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr12_+_8309630 | 0.05 |
ENST00000396570.3
|
ZNF705A
|
zinc finger protein 705A |
| chr2_-_287320 | 0.05 |
ENST00000401503.1
|
FAM150B
|
family with sequence similarity 150, member B |
| chr1_-_76398077 | 0.04 |
ENST00000284142.6
|
ASB17
|
ankyrin repeat and SOCS box containing 17 |
| chr17_-_50236039 | 0.04 |
ENST00000451037.2
|
CA10
|
carbonic anhydrase X |
| chr3_-_123680246 | 0.04 |
ENST00000488653.2
|
CCDC14
|
coiled-coil domain containing 14 |
| chr2_+_1417228 | 0.04 |
ENST00000382269.3
ENST00000337415.3 ENST00000345913.4 ENST00000346956.3 ENST00000349624.3 ENST00000539820.1 ENST00000329066.4 ENST00000382201.3 |
TPO
|
thyroid peroxidase |
| chr6_+_106535455 | 0.04 |
ENST00000424894.1
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr17_-_4607335 | 0.03 |
ENST00000570571.1
ENST00000575101.1 ENST00000436683.2 ENST00000574876.1 |
PELP1
|
proline, glutamate and leucine rich protein 1 |
| chr2_+_177134201 | 0.03 |
ENST00000452865.1
|
MTX2
|
metaxin 2 |
| chr2_-_130031335 | 0.03 |
ENST00000375987.3
|
AC079586.1
|
AC079586.1 |
| chr1_+_197886461 | 0.03 |
ENST00000367388.3
ENST00000337020.2 ENST00000367387.4 |
LHX9
|
LIM homeobox 9 |
| chrX_-_23926004 | 0.03 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr15_+_88120158 | 0.02 |
ENST00000560153.1
|
LINC00052
|
long intergenic non-protein coding RNA 52 |
| chr9_+_2157655 | 0.01 |
ENST00000452193.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_-_157221128 | 0.01 |
ENST00000392833.2
ENST00000362010.2 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr18_-_29340827 | 0.01 |
ENST00000269205.5
|
SLC25A52
|
solute carrier family 25, member 52 |
| chr14_-_65409502 | 0.01 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chrX_-_77150985 | 0.00 |
ENST00000358075.6
|
MAGT1
|
magnesium transporter 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HMGA2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of auditory receptor cell differentiation(GO:0045608) regulation of timing of neuron differentiation(GO:0060164) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.2 | 0.7 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.1 | 0.4 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 0.4 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 | 0.9 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.3 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 2.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.8 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 0.2 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 0.3 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.4 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.2 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) beta-alanine metabolic process(GO:0019482) thymine metabolic process(GO:0019859) uracil metabolic process(GO:0019860) |
| 0.0 | 0.7 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.2 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.2 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.2 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 0.1 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.9 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.2 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0070779 | gamma-aminobutyric acid biosynthetic process(GO:0009449) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.2 | GO:0072564 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.0 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.2 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 0.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 2.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.2 | 0.8 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.7 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 2.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.3 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0002054 | nucleobase binding(GO:0002054) dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.8 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 2.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.7 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.7 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |