Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
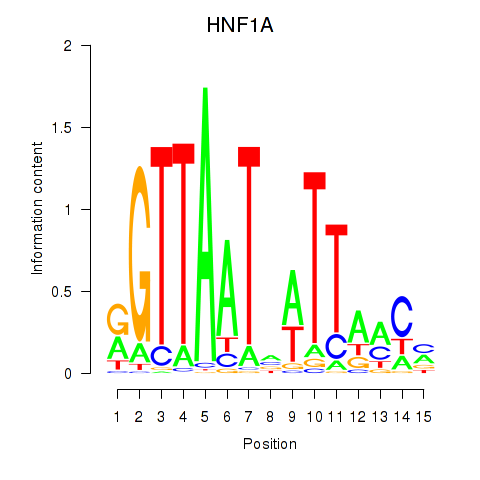
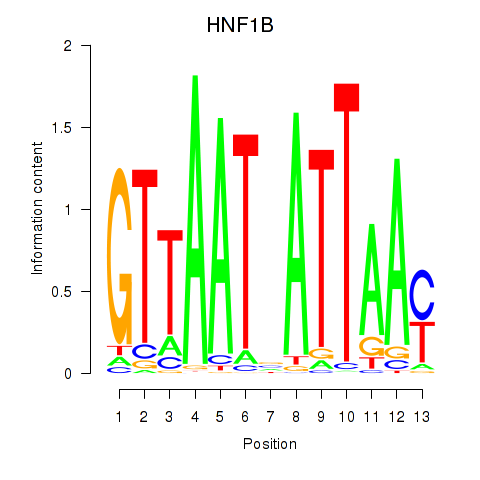
Results for HNF1A_HNF1B
Z-value: 5.64
Transcription factors associated with HNF1A_HNF1B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HNF1A
|
ENSG00000135100.13 | HNF1 homeobox A |
|
HNF1B
|
ENSG00000108753.8 | HNF1 homeobox B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HNF1A | hg19_v2_chr12_+_121416340_121416371 | 0.94 | 5.9e-10 | Click! |
| HNF1B | hg19_v2_chr17_-_36105009_36105060 | 0.82 | 1.1e-05 | Click! |
Activity profile of HNF1A_HNF1B motif
Sorted Z-values of HNF1A_HNF1B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_64225508 | 46.77 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr10_+_101542462 | 41.10 |
ENST00000370449.4
ENST00000370434.1 |
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr4_-_155511887 | 40.74 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr14_-_57960456 | 37.93 |
ENST00000534126.1
ENST00000422976.2 |
C14orf105
|
chromosome 14 open reading frame 105 |
| chr9_-_123812542 | 36.87 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr14_-_57960545 | 36.84 |
ENST00000526336.1
ENST00000216445.3 |
C14orf105
|
chromosome 14 open reading frame 105 |
| chr5_+_176513895 | 36.50 |
ENST00000503708.1
ENST00000393648.2 ENST00000514472.1 ENST00000502906.1 ENST00000292410.3 ENST00000510911.1 |
FGFR4
|
fibroblast growth factor receptor 4 |
| chr19_-_36304201 | 36.39 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr4_+_155484155 | 35.51 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr4_+_155484103 | 35.08 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr22_-_30970560 | 34.95 |
ENST00000402369.1
ENST00000406361.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr8_-_124749609 | 32.92 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr20_+_43029911 | 32.69 |
ENST00000443598.2
ENST00000316099.4 ENST00000415691.2 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr17_+_73642315 | 30.08 |
ENST00000556126.2
|
SMIM6
|
small integral membrane protein 6 |
| chr11_-_117695449 | 28.06 |
ENST00000292079.2
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr17_+_73642486 | 27.69 |
ENST00000579469.1
|
SMIM6
|
small integral membrane protein 6 |
| chr10_+_5135981 | 27.60 |
ENST00000380554.3
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr5_+_176513868 | 27.56 |
ENST00000292408.4
|
FGFR4
|
fibroblast growth factor receptor 4 |
| chr22_-_30968813 | 27.34 |
ENST00000443111.2
ENST00000443136.1 ENST00000426220.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr7_+_45927956 | 26.51 |
ENST00000275525.3
ENST00000457280.1 |
IGFBP1
|
insulin-like growth factor binding protein 1 |
| chr20_+_42187682 | 22.98 |
ENST00000373092.3
ENST00000373077.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr22_-_30968839 | 22.76 |
ENST00000445645.1
ENST00000416358.1 ENST00000423371.1 ENST00000411821.1 ENST00000448604.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr16_+_81272287 | 22.51 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr2_-_228244013 | 22.20 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr22_-_30970498 | 20.81 |
ENST00000431313.1
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr7_+_45928079 | 20.57 |
ENST00000468955.1
|
IGFBP1
|
insulin-like growth factor binding protein 1 |
| chr8_-_95220775 | 19.83 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr9_-_116861337 | 19.43 |
ENST00000374118.3
|
KIF12
|
kinesin family member 12 |
| chr8_-_17752996 | 19.08 |
ENST00000381841.2
ENST00000427924.1 |
FGL1
|
fibrinogen-like 1 |
| chr1_+_207277590 | 18.15 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr16_+_20775024 | 17.46 |
ENST00000289416.5
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr1_-_204183071 | 17.34 |
ENST00000308302.3
|
GOLT1A
|
golgi transport 1A |
| chr17_-_202579 | 17.04 |
ENST00000577079.1
ENST00000331302.7 ENST00000536489.2 |
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr17_+_4692230 | 16.79 |
ENST00000331264.7
|
GLTPD2
|
glycolipid transfer protein domain containing 2 |
| chr7_-_27219849 | 16.79 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr16_+_20775358 | 16.16 |
ENST00000440284.2
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr16_-_57831914 | 15.69 |
ENST00000421376.2
|
KIFC3
|
kinesin family member C3 |
| chr5_+_176514413 | 15.61 |
ENST00000513166.1
|
FGFR4
|
fibroblast growth factor receptor 4 |
| chr20_+_42984330 | 15.60 |
ENST00000316673.4
ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr22_-_38506619 | 15.40 |
ENST00000332536.5
ENST00000381669.3 |
BAIAP2L2
|
BAI1-associated protein 2-like 2 |
| chr20_+_42187608 | 14.63 |
ENST00000373100.1
|
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr5_-_138718973 | 13.90 |
ENST00000353963.3
ENST00000348729.3 |
SLC23A1
|
solute carrier family 23 (ascorbic acid transporter), member 1 |
| chr3_+_183948161 | 13.42 |
ENST00000426955.2
|
VWA5B2
|
von Willebrand factor A domain containing 5B2 |
| chr19_+_50016610 | 12.52 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr14_+_21156915 | 11.65 |
ENST00000397990.4
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr1_+_18807424 | 11.11 |
ENST00000400664.1
|
KLHDC7A
|
kelch domain containing 7A |
| chr12_-_8693469 | 10.32 |
ENST00000545274.1
ENST00000446457.2 |
CLEC4E
|
C-type lectin domain family 4, member E |
| chr7_-_50633078 | 10.24 |
ENST00000444124.2
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr6_-_25874440 | 10.18 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chr12_+_20963632 | 9.90 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr2_-_47572207 | 9.70 |
ENST00000441997.1
|
AC073283.4
|
AC073283.4 |
| chr2_-_99917639 | 9.57 |
ENST00000308528.4
|
LYG1
|
lysozyme G-like 1 |
| chr11_+_560956 | 9.40 |
ENST00000397582.3
ENST00000344375.4 ENST00000397583.3 |
RASSF7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr10_+_115312766 | 9.30 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chr2_-_47572105 | 9.26 |
ENST00000419035.1
ENST00000448713.1 ENST00000450550.1 ENST00000413185.2 |
AC073283.4
|
AC073283.4 |
| chr17_-_26733179 | 9.21 |
ENST00000440501.1
ENST00000321666.5 |
SLC46A1
|
solute carrier family 46 (folate transporter), member 1 |
| chr16_-_57831676 | 9.15 |
ENST00000465878.2
ENST00000539578.1 ENST00000561524.1 |
KIFC3
|
kinesin family member C3 |
| chr14_-_54425475 | 9.03 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr12_-_8693539 | 8.54 |
ENST00000299663.3
|
CLEC4E
|
C-type lectin domain family 4, member E |
| chr10_+_99344071 | 8.48 |
ENST00000370647.4
ENST00000370646.4 |
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr12_+_20963647 | 8.45 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr2_+_175199674 | 8.39 |
ENST00000394967.2
|
SP9
|
Sp9 transcription factor |
| chr19_+_54466179 | 8.39 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr18_-_70931689 | 8.31 |
ENST00000581862.1
|
RP11-169F17.1
|
Protein LOC400655 |
| chr6_+_31707725 | 8.29 |
ENST00000375755.3
ENST00000375742.3 ENST00000375750.3 ENST00000425703.1 ENST00000534153.4 ENST00000375703.3 ENST00000375740.3 |
MSH5
|
mutS homolog 5 |
| chr2_+_133174147 | 8.20 |
ENST00000329321.3
|
GPR39
|
G protein-coupled receptor 39 |
| chr11_-_64660916 | 8.02 |
ENST00000413053.1
|
MIR194-2
|
microRNA 194-2 |
| chr20_+_35973080 | 7.88 |
ENST00000445403.1
|
SRC
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr19_+_50016411 | 7.84 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr17_-_26733604 | 7.46 |
ENST00000584426.1
ENST00000584995.1 |
SLC46A1
|
solute carrier family 46 (folate transporter), member 1 |
| chr11_-_65149422 | 7.39 |
ENST00000526432.1
ENST00000527174.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chr1_+_207277632 | 7.27 |
ENST00000421786.1
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr3_-_137834436 | 7.08 |
ENST00000327532.2
ENST00000467030.1 |
DZIP1L
|
DAZ interacting zinc finger protein 1-like |
| chr16_+_21244986 | 6.95 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr4_-_987217 | 6.53 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr16_-_57832004 | 6.44 |
ENST00000562503.1
|
KIFC3
|
kinesin family member C3 |
| chr14_-_65409502 | 6.21 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr7_-_47579188 | 5.99 |
ENST00000398879.1
ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3
|
tensin 3 |
| chr2_-_87248975 | 5.94 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr7_-_155601766 | 5.92 |
ENST00000430104.1
|
SHH
|
sonic hedgehog |
| chr6_-_127840453 | 5.85 |
ENST00000556132.1
|
SOGA3
|
SOGA family member 3 |
| chr19_-_57678811 | 5.59 |
ENST00000554048.2
|
DUXA
|
double homeobox A |
| chr8_+_21911054 | 5.52 |
ENST00000519850.1
ENST00000381470.3 |
DMTN
|
dematin actin binding protein |
| chr6_+_126221034 | 5.46 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr1_+_55505184 | 5.35 |
ENST00000302118.5
|
PCSK9
|
proprotein convertase subtilisin/kexin type 9 |
| chr10_+_115312825 | 5.33 |
ENST00000537906.1
ENST00000541666.1 |
HABP2
|
hyaluronan binding protein 2 |
| chr6_-_127840021 | 5.07 |
ENST00000465909.2
|
SOGA3
|
SOGA family member 3 |
| chr6_-_127840336 | 5.01 |
ENST00000525778.1
|
SOGA3
|
SOGA family member 3 |
| chr14_+_39703112 | 4.89 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr6_+_161123270 | 4.83 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr17_+_79953310 | 4.74 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr4_-_110723194 | 4.57 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr11_+_46740730 | 4.53 |
ENST00000311907.5
ENST00000530231.1 ENST00000442468.1 |
F2
|
coagulation factor II (thrombin) |
| chr2_+_38177575 | 4.47 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr1_-_114430169 | 4.42 |
ENST00000393316.3
|
BCL2L15
|
BCL2-like 15 |
| chr12_+_100867694 | 4.39 |
ENST00000392986.3
ENST00000549996.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr16_-_66952779 | 4.26 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr7_+_141695633 | 4.16 |
ENST00000549489.2
|
MGAM
|
maltase-glucoamylase (alpha-glucosidase) |
| chr9_-_94712434 | 4.08 |
ENST00000375708.3
|
ROR2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr14_-_65409438 | 4.06 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr2_-_55920952 | 3.93 |
ENST00000447944.2
|
PNPT1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr4_-_110723335 | 3.91 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr4_-_987164 | 3.91 |
ENST00000398520.2
|
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr9_-_98079965 | 3.83 |
ENST00000289081.3
|
FANCC
|
Fanconi anemia, complementation group C |
| chr7_+_130126012 | 3.82 |
ENST00000341441.5
|
MEST
|
mesoderm specific transcript |
| chr12_+_100867486 | 3.82 |
ENST00000548884.1
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr20_+_62371206 | 3.68 |
ENST00000266077.2
|
SLC2A4RG
|
SLC2A4 regulator |
| chr7_+_141695671 | 3.59 |
ENST00000497673.1
ENST00000475668.2 |
MGAM
|
maltase-glucoamylase (alpha-glucosidase) |
| chr6_-_127840048 | 3.54 |
ENST00000467753.1
|
SOGA3
|
SOGA family member 3 |
| chr1_+_48688357 | 3.52 |
ENST00000533824.1
ENST00000438567.2 ENST00000236495.5 ENST00000420136.2 |
SLC5A9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr22_+_31003190 | 3.49 |
ENST00000407817.3
|
TCN2
|
transcobalamin II |
| chr4_+_3443614 | 3.48 |
ENST00000382774.3
ENST00000511533.1 |
HGFAC
|
HGF activator |
| chr16_-_66952742 | 3.44 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr3_-_157221380 | 3.44 |
ENST00000468233.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr11_+_64323156 | 3.42 |
ENST00000377585.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr7_-_27219632 | 3.24 |
ENST00000470747.4
|
RP1-170O19.20
|
Uncharacterized protein |
| chr11_+_64323098 | 3.22 |
ENST00000301891.4
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr2_+_27719697 | 3.21 |
ENST00000264717.2
ENST00000424318.2 |
GCKR
|
glucokinase (hexokinase 4) regulator |
| chr16_-_420514 | 3.20 |
ENST00000199706.8
|
MRPL28
|
mitochondrial ribosomal protein L28 |
| chr22_+_31003133 | 3.19 |
ENST00000405742.3
|
TCN2
|
transcobalamin II |
| chr7_+_130126165 | 3.19 |
ENST00000427521.1
ENST00000416162.2 ENST00000378576.4 |
MEST
|
mesoderm specific transcript |
| chr12_+_100897130 | 3.17 |
ENST00000551379.1
ENST00000188403.7 ENST00000551184.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr6_-_161085291 | 3.16 |
ENST00000316300.5
|
LPA
|
lipoprotein, Lp(a) |
| chr10_-_128359074 | 3.08 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr10_-_52645416 | 3.04 |
ENST00000374001.2
ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr22_+_31002779 | 2.98 |
ENST00000215838.3
|
TCN2
|
transcobalamin II |
| chr3_-_157221128 | 2.98 |
ENST00000392833.2
ENST00000362010.2 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr22_-_32651326 | 2.96 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr1_+_207262881 | 2.81 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr19_-_11639910 | 2.77 |
ENST00000588998.1
ENST00000586149.1 |
ECSIT
|
ECSIT signalling integrator |
| chr21_-_19775973 | 2.72 |
ENST00000284885.3
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr3_-_157221357 | 2.64 |
ENST00000494677.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr10_-_72648541 | 2.58 |
ENST00000299299.3
|
PCBD1
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
| chr17_+_68071389 | 2.57 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr20_-_7921090 | 2.49 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr7_-_47578840 | 2.49 |
ENST00000450444.1
|
TNS3
|
tensin 3 |
| chr6_+_25754927 | 2.48 |
ENST00000377905.4
ENST00000439485.2 |
SLC17A4
|
solute carrier family 17, member 4 |
| chr6_-_52628271 | 2.47 |
ENST00000493422.1
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr12_+_21284118 | 2.42 |
ENST00000256958.2
|
SLCO1B1
|
solute carrier organic anion transporter family, member 1B1 |
| chr4_+_124571409 | 2.39 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr5_-_77844974 | 2.38 |
ENST00000515007.2
|
LHFPL2
|
lipoma HMGIC fusion partner-like 2 |
| chr21_-_35340759 | 2.36 |
ENST00000607953.1
|
AP000569.9
|
AP000569.9 |
| chrX_+_120181457 | 2.33 |
ENST00000328078.1
|
GLUD2
|
glutamate dehydrogenase 2 |
| chr12_+_100867733 | 2.33 |
ENST00000546380.1
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr8_-_108510224 | 2.32 |
ENST00000517746.1
ENST00000297450.3 |
ANGPT1
|
angiopoietin 1 |
| chr6_+_54173227 | 2.30 |
ENST00000259782.4
ENST00000370864.3 |
TINAG
|
tubulointerstitial nephritis antigen |
| chr6_-_116833500 | 2.30 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr3_-_107596910 | 2.29 |
ENST00000464359.2
ENST00000464823.1 ENST00000466155.1 ENST00000473528.2 ENST00000608306.1 ENST00000488852.1 ENST00000608137.1 ENST00000608307.1 ENST00000609429.1 ENST00000601385.1 ENST00000475362.1 ENST00000600240.1 ENST00000600749.1 |
LINC00635
|
long intergenic non-protein coding RNA 635 |
| chr4_+_74269956 | 2.29 |
ENST00000295897.4
ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB
|
albumin |
| chr11_+_20620946 | 2.25 |
ENST00000525748.1
|
SLC6A5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr9_+_140125385 | 2.25 |
ENST00000361134.2
|
SLC34A3
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 3 |
| chr5_+_175976324 | 2.18 |
ENST00000261944.5
|
CDHR2
|
cadherin-related family member 2 |
| chr1_+_66999799 | 2.16 |
ENST00000371035.3
ENST00000371036.3 ENST00000371037.4 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr19_-_11639931 | 2.12 |
ENST00000592312.1
ENST00000590480.1 ENST00000585318.1 ENST00000252440.7 ENST00000417981.2 ENST00000270517.7 |
ECSIT
|
ECSIT signalling integrator |
| chr9_+_130860810 | 2.12 |
ENST00000433501.1
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr3_-_192445289 | 2.07 |
ENST00000430714.1
ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12
|
fibroblast growth factor 12 |
| chr14_+_39703084 | 2.00 |
ENST00000553728.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr12_-_102874416 | 1.99 |
ENST00000392904.1
ENST00000337514.6 |
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr16_+_22524379 | 1.98 |
ENST00000536620.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr17_+_68071458 | 1.84 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr4_+_14113592 | 1.78 |
ENST00000502759.1
ENST00000511200.1 ENST00000512754.1 ENST00000506739.1 |
LINC01085
|
long intergenic non-protein coding RNA 1085 |
| chr17_-_36904437 | 1.76 |
ENST00000585100.1
ENST00000360797.2 ENST00000578109.1 ENST00000579882.1 |
PCGF2
|
polycomb group ring finger 2 |
| chr1_+_84767289 | 1.73 |
ENST00000394834.3
ENST00000370669.1 |
SAMD13
|
sterile alpha motif domain containing 13 |
| chr22_+_31002990 | 1.67 |
ENST00000423350.1
|
TCN2
|
transcobalamin II |
| chr9_+_130860583 | 1.66 |
ENST00000373064.5
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chrX_-_124097620 | 1.63 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr17_+_1646130 | 1.62 |
ENST00000453066.1
ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr3_-_169587621 | 1.61 |
ENST00000523069.1
ENST00000316428.5 ENST00000264676.5 |
LRRC31
|
leucine rich repeat containing 31 |
| chr19_-_43099070 | 1.57 |
ENST00000244336.5
|
CEACAM8
|
carcinoembryonic antigen-related cell adhesion molecule 8 |
| chr7_+_120629653 | 1.50 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr2_-_158300556 | 1.48 |
ENST00000264192.3
|
CYTIP
|
cytohesin 1 interacting protein |
| chr3_-_49466686 | 1.47 |
ENST00000273598.3
ENST00000436744.2 |
NICN1
|
nicolin 1 |
| chr19_+_41305085 | 1.44 |
ENST00000303961.4
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr15_+_58430567 | 1.40 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr4_-_70518941 | 1.40 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr8_+_55047763 | 1.39 |
ENST00000260102.4
ENST00000519831.1 |
MRPL15
|
mitochondrial ribosomal protein L15 |
| chr3_-_161089289 | 1.33 |
ENST00000497137.1
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chr15_+_58430368 | 1.32 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr6_+_167704838 | 1.31 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr16_-_20587599 | 1.28 |
ENST00000566384.1
ENST00000565232.1 ENST00000567001.1 ENST00000565322.1 ENST00000569344.1 ENST00000329697.6 ENST00000414188.2 ENST00000568882.1 |
ACSM2B
|
acyl-CoA synthetase medium-chain family member 2B |
| chrX_+_36254051 | 1.20 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr17_+_48823975 | 1.20 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr9_-_215744 | 1.19 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr16_-_55909211 | 1.15 |
ENST00000520435.1
|
CES5A
|
carboxylesterase 5A |
| chr1_-_159684371 | 1.15 |
ENST00000255030.5
ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP
|
C-reactive protein, pentraxin-related |
| chr12_-_54653313 | 1.15 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr1_-_197036364 | 1.11 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr4_+_141264597 | 1.10 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr1_+_17906970 | 1.09 |
ENST00000375415.1
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr17_+_48823896 | 1.02 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr16_-_55909255 | 1.00 |
ENST00000290567.9
|
CES5A
|
carboxylesterase 5A |
| chr16_-_55909272 | 0.99 |
ENST00000319165.9
|
CES5A
|
carboxylesterase 5A |
| chr10_-_38146510 | 0.97 |
ENST00000395867.3
|
ZNF248
|
zinc finger protein 248 |
| chr14_+_24590560 | 0.96 |
ENST00000558325.1
|
RP11-468E2.6
|
RP11-468E2.6 |
| chr6_+_54172653 | 0.96 |
ENST00000370869.3
|
TINAG
|
tubulointerstitial nephritis antigen |
| chr13_+_96085847 | 0.95 |
ENST00000376873.3
|
CLDN10
|
claudin 10 |
| chr10_-_101690650 | 0.93 |
ENST00000543621.1
|
DNMBP
|
dynamin binding protein |
| chr1_+_207262578 | 0.92 |
ENST00000243611.5
ENST00000367076.3 |
C4BPB
|
complement component 4 binding protein, beta |
| chr9_+_140125209 | 0.92 |
ENST00000538474.1
|
SLC34A3
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 3 |
| chr12_-_102874102 | 0.89 |
ENST00000392905.2
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
Network of associatons between targets according to the STRING database.
First level regulatory network of HNF1A_HNF1B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.0 | 41.9 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 13.3 | 79.7 | GO:1903412 | response to bile acid(GO:1903412) |
| 9.2 | 27.6 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 8.1 | 105.9 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 7.9 | 110.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 6.8 | 20.4 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 6.1 | 36.9 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 5.2 | 36.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 4.8 | 48.3 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 4.6 | 13.9 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 4.3 | 51.4 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 4.2 | 8.5 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 4.0 | 19.8 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 3.4 | 13.7 | GO:2000213 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 3.4 | 10.2 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 3.1 | 31.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 3.1 | 30.7 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 2.8 | 16.7 | GO:0051958 | methotrexate transport(GO:0051958) |
| 2.0 | 5.9 | GO:0060738 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of immature T cell proliferation in thymus(GO:0033092) right lung development(GO:0060458) primary prostatic bud elongation(GO:0060516) epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 2.0 | 7.9 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 1.9 | 11.7 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.8 | 9.0 | GO:2000005 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 1.8 | 5.3 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 1.7 | 7.0 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 1.6 | 4.8 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 1.5 | 4.5 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 1.4 | 28.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 1.4 | 47.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 1.4 | 14.9 | GO:0015747 | urate transport(GO:0015747) |
| 1.3 | 3.9 | GO:2000627 | rRNA import into mitochondrion(GO:0035928) regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 1.2 | 9.6 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 1.2 | 7.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 1.0 | 10.4 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.9 | 7.4 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.9 | 11.3 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.8 | 2.5 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.8 | 2.3 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.8 | 8.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.7 | 2.2 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.7 | 4.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.6 | 20.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.6 | 15.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.6 | 5.5 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.5 | 2.7 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.5 | 3.2 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.5 | 2.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.5 | 16.8 | GO:0060065 | uterus development(GO:0060065) |
| 0.4 | 1.6 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.4 | 3.2 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.3 | 32.1 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.3 | 3.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.3 | 17.0 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.3 | 3.2 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.3 | 8.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.3 | 5.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.3 | 2.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.3 | 36.5 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.2 | 2.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 3.8 | GO:0015866 | ADP transport(GO:0015866) |
| 0.2 | 2.6 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.2 | 2.3 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.2 | 2.3 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.2 | 8.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 1.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.2 | 1.3 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.2 | 8.5 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.2 | 18.9 | GO:0002292 | T cell differentiation involved in immune response(GO:0002292) |
| 0.1 | 0.6 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.1 | 0.7 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 | 4.7 | GO:0010827 | regulation of glucose transport(GO:0010827) regulation of glucose import(GO:0046324) |
| 0.1 | 0.7 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.1 | 0.4 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.1 | 1.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 4.9 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.4 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.1 | 1.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 9.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 14.6 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.1 | 2.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 1.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 1.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.7 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 1.1 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 4.9 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.5 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.5 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 5.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 8.4 | GO:0035113 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.0 | 18.1 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.5 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 1.6 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 2.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.5 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 9.9 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 6.6 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 0.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.8 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 0.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 13.1 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 7.1 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.8 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 2.3 | GO:0007599 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.0 | 0.3 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 2.4 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.0 | 0.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 2.4 | GO:0007188 | adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.0 | 5.1 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.2 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 4.9 | GO:0045861 | negative regulation of proteolysis(GO:0045861) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.4 | 132.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 4.6 | 36.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 3.3 | 39.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 2.6 | 46.7 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 1.8 | 5.3 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 1.7 | 11.7 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 1.3 | 3.8 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 1.3 | 31.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 1.1 | 28.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.9 | 15.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.7 | 4.8 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.7 | 4.6 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.5 | 8.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.4 | 2.2 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.4 | 4.1 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.3 | 3.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.3 | 3.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 13.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.2 | 5.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 13.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 30.7 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.2 | 18.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 18.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 8.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 4.7 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 1.8 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 4.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 79.3 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.1 | 2.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 48.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 2.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 7.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 2.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 1.3 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 11.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 105.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 40.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 9.3 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 58.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 2.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 1.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 29.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 11.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 3.2 | GO:0034358 | plasma lipoprotein particle(GO:0034358) lipoprotein particle(GO:1990777) |
| 0.1 | 7.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 1.9 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 3.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 2.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 3.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 21.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 16.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.9 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 5.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 27.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 7.5 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 26.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 1.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 47.4 | GO:0005654 | nucleoplasm(GO:0005654) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.4 | 46.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 9.2 | 27.6 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 9.1 | 36.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 8.8 | 105.9 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 6.6 | 79.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 6.6 | 19.8 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 4.6 | 13.9 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 4.1 | 20.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 3.4 | 13.7 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 3.4 | 47.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 3.2 | 35.6 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 2.8 | 16.7 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 2.2 | 41.1 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 1.7 | 11.7 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 1.6 | 4.8 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 1.4 | 17.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.3 | 37.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 1.3 | 5.3 | GO:0070326 | very-low-density lipoprotein particle binding(GO:0034189) very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 1.3 | 28.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 1.3 | 14.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 1.2 | 8.5 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 1.1 | 7.4 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 1.0 | 7.3 | GO:0016160 | amylase activity(GO:0016160) |
| 1.0 | 31.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.9 | 2.7 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.9 | 4.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.9 | 11.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.9 | 2.6 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.9 | 2.6 | GO:0008124 | phenylalanine 4-monooxygenase activity(GO:0004505) 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.8 | 36.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.8 | 9.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.8 | 46.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.7 | 18.6 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.7 | 5.9 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.7 | 7.9 | GO:0071253 | connexin binding(GO:0071253) |
| 0.6 | 9.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.6 | 3.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.6 | 2.3 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.6 | 2.3 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.6 | 3.9 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.5 | 10.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.5 | 4.9 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.5 | 4.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.4 | 70.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.4 | 8.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.3 | 10.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.3 | 4.5 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.3 | 5.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.3 | 4.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.3 | 3.8 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 0.8 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.2 | 1.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.2 | 0.7 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.2 | 25.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 14.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 1.3 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 18.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 10.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 0.7 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 8.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 1.6 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 1.4 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 1.8 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 18.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 5.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.5 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.4 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.8 | GO:0034235 | GPI anchor binding(GO:0034235) metallodipeptidase activity(GO:0070573) |
| 0.1 | 6.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.4 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.1 | 24.7 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 17.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.5 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 0.9 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 18.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 2.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 3.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 4.9 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.6 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 7.0 | GO:0016787 | hydrolase activity(GO:0016787) |
| 0.0 | 3.7 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 11.4 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 4.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 9.2 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 8.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 113.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.9 | 83.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.7 | 79.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.4 | 25.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.4 | 16.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 29.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 13.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 16.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 35.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 8.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 9.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 4.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 29.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 5.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 3.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 4.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 4.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 4.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 117.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 4.7 | 79.7 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 2.0 | 51.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 1.5 | 78.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 1.5 | 105.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 1.2 | 20.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 1.1 | 48.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.8 | 41.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.6 | 31.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.4 | 28.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.4 | 22.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.4 | 10.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 7.9 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.3 | 4.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.3 | 30.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 4.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.2 | 2.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 2.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 3.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 8.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 12.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 3.2 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 2.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 7.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.7 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 1.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 4.9 | REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.0 | 9.8 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 3.8 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 1.2 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |