Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for HNF4A
Z-value: 2.00
Transcription factors associated with HNF4A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HNF4A
|
ENSG00000101076.12 | hepatocyte nuclear factor 4 alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HNF4A | hg19_v2_chr20_+_43029911_43029941 | 0.94 | 7.5e-10 | Click! |
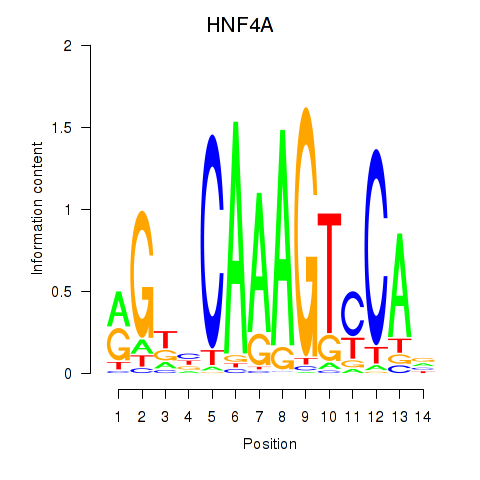
Activity profile of HNF4A motif
Sorted Z-values of HNF4A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_73606766 | 18.63 |
ENST00000578462.1
|
MYO15B
|
myosin XVB pseudogene |
| chr2_-_228244013 | 11.66 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr17_-_36105009 | 10.98 |
ENST00000560016.1
ENST00000427275.2 ENST00000561193.1 |
HNF1B
|
HNF1 homeobox B |
| chr3_-_120400960 | 9.52 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr3_-_124653579 | 9.40 |
ENST00000478191.1
ENST00000311075.3 |
MUC13
|
mucin 13, cell surface associated |
| chr12_+_56075330 | 8.43 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr7_+_45927956 | 7.92 |
ENST00000275525.3
ENST00000457280.1 |
IGFBP1
|
insulin-like growth factor binding protein 1 |
| chr16_+_81272287 | 6.29 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr12_+_47617284 | 6.25 |
ENST00000549630.1
ENST00000551777.1 |
PCED1B
|
PC-esterase domain containing 1B |
| chr7_+_45928079 | 6.08 |
ENST00000468955.1
|
IGFBP1
|
insulin-like growth factor binding protein 1 |
| chrX_+_66764375 | 6.04 |
ENST00000374690.3
|
AR
|
androgen receptor |
| chr1_+_199996702 | 6.01 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr12_+_121416489 | 5.46 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr6_-_30080876 | 5.18 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr6_-_30080863 | 5.17 |
ENST00000540829.1
|
TRIM31
|
tripartite motif containing 31 |
| chr12_+_121416340 | 5.10 |
ENST00000257555.6
ENST00000400024.2 |
HNF1A
|
HNF1 homeobox A |
| chr16_-_16317321 | 4.95 |
ENST00000205557.7
ENST00000575728.1 |
ABCC6
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6 |
| chr19_+_39616410 | 4.79 |
ENST00000602004.1
ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr17_+_4853442 | 4.77 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr2_+_241544834 | 4.75 |
ENST00000319838.5
ENST00000403859.1 ENST00000438013.2 |
GPR35
|
G protein-coupled receptor 35 |
| chr17_+_4675175 | 4.44 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr2_+_10442993 | 4.28 |
ENST00000423674.1
ENST00000307845.3 |
HPCAL1
|
hippocalcin-like 1 |
| chr1_+_199996733 | 3.99 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr19_-_36304201 | 3.91 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr16_-_57514277 | 3.80 |
ENST00000562008.1
ENST00000567214.1 |
DOK4
|
docking protein 4 |
| chr22_+_25003626 | 3.76 |
ENST00000451366.1
ENST00000406383.2 ENST00000428855.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr7_-_37956409 | 3.58 |
ENST00000436072.2
|
SFRP4
|
secreted frizzled-related protein 4 |
| chr19_+_18496957 | 3.52 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr11_-_65381643 | 3.32 |
ENST00000309100.3
ENST00000529839.1 ENST00000526293.1 |
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr12_+_56114151 | 3.29 |
ENST00000547072.1
ENST00000552930.1 ENST00000257895.5 |
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr5_+_133842243 | 3.28 |
ENST00000515627.2
|
AC005355.2
|
AC005355.2 |
| chr15_-_63448973 | 3.24 |
ENST00000462430.1
|
RPS27L
|
ribosomal protein S27-like |
| chr7_+_192969 | 3.22 |
ENST00000313766.5
|
FAM20C
|
family with sequence similarity 20, member C |
| chr7_-_15601595 | 3.11 |
ENST00000342526.3
|
AGMO
|
alkylglycerol monooxygenase |
| chr17_-_44896047 | 3.10 |
ENST00000225512.5
|
WNT3
|
wingless-type MMTV integration site family, member 3 |
| chr4_+_95376396 | 3.07 |
ENST00000508216.1
ENST00000514743.1 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr17_-_17494972 | 2.99 |
ENST00000435340.2
ENST00000255389.5 ENST00000395781.2 |
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr12_+_121416437 | 2.97 |
ENST00000402929.1
ENST00000535955.1 ENST00000538626.1 ENST00000543427.1 |
HNF1A
|
HNF1 homeobox A |
| chr12_+_121163538 | 2.96 |
ENST00000242592.4
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr11_+_119056178 | 2.95 |
ENST00000525131.1
ENST00000531114.1 ENST00000355547.5 ENST00000322712.4 |
PDZD3
|
PDZ domain containing 3 |
| chr19_-_49339080 | 2.84 |
ENST00000595764.1
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr7_-_47621229 | 2.80 |
ENST00000434451.1
|
TNS3
|
tensin 3 |
| chr2_+_74648848 | 2.76 |
ENST00000409791.1
ENST00000426787.1 ENST00000348227.4 |
WDR54
|
WD repeat domain 54 |
| chr12_+_56114189 | 2.74 |
ENST00000548082.1
|
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr22_+_25003606 | 2.73 |
ENST00000432867.1
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr17_+_4692230 | 2.73 |
ENST00000331264.7
|
GLTPD2
|
glycolipid transfer protein domain containing 2 |
| chr10_+_96698406 | 2.61 |
ENST00000260682.6
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr22_+_25003568 | 2.59 |
ENST00000447416.1
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr2_-_28113217 | 2.57 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr2_-_74645669 | 2.56 |
ENST00000518401.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr17_+_7591747 | 2.55 |
ENST00000534050.1
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr1_-_177939041 | 2.53 |
ENST00000308284.6
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr17_-_7082861 | 2.51 |
ENST00000269299.3
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr5_-_180235755 | 2.49 |
ENST00000502678.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr12_+_121163602 | 2.48 |
ENST00000411593.2
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chrX_+_118108571 | 2.47 |
ENST00000304778.7
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr14_+_32476072 | 2.45 |
ENST00000556949.1
|
RP11-187E13.2
|
Uncharacterized protein |
| chr2_-_74648702 | 2.36 |
ENST00000518863.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr2_+_28113583 | 2.34 |
ENST00000344773.2
ENST00000379624.1 ENST00000342045.2 ENST00000379632.2 ENST00000361704.2 |
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chr17_+_7591639 | 2.34 |
ENST00000396463.2
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr3_-_53878644 | 2.28 |
ENST00000481668.1
ENST00000467802.1 |
CHDH
|
choline dehydrogenase |
| chr19_-_10687983 | 2.27 |
ENST00000587069.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr19_+_49055332 | 2.26 |
ENST00000201586.2
|
SULT2B1
|
sulfotransferase family, cytosolic, 2B, member 1 |
| chr20_+_57875457 | 2.16 |
ENST00000337938.2
ENST00000311585.7 ENST00000371028.2 |
EDN3
|
endothelin 3 |
| chr19_-_41256207 | 2.15 |
ENST00000598485.2
ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr11_-_73720122 | 2.15 |
ENST00000426995.2
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr2_+_219135115 | 2.10 |
ENST00000248451.3
ENST00000273077.4 |
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr11_-_73720276 | 2.05 |
ENST00000348534.4
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr20_+_62694461 | 2.04 |
ENST00000343484.5
ENST00000395053.3 |
TCEA2
|
transcription elongation factor A (SII), 2 |
| chr19_-_48867171 | 2.03 |
ENST00000377431.2
ENST00000436660.2 ENST00000541566.1 |
TMEM143
|
transmembrane protein 143 |
| chr5_-_79950371 | 2.02 |
ENST00000511032.1
ENST00000504396.1 ENST00000505337.1 |
DHFR
|
dihydrofolate reductase |
| chrX_+_118108601 | 2.01 |
ENST00000371628.3
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr2_+_73441350 | 1.90 |
ENST00000389501.4
|
SMYD5
|
SMYD family member 5 |
| chr4_-_120243545 | 1.89 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr13_+_113777105 | 1.88 |
ENST00000409306.1
ENST00000375551.3 ENST00000375559.3 |
F10
|
coagulation factor X |
| chr11_-_45939374 | 1.84 |
ENST00000533151.1
ENST00000241041.3 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr19_-_10687948 | 1.82 |
ENST00000592285.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr16_+_24857552 | 1.76 |
ENST00000568579.1
ENST00000567758.1 ENST00000569071.1 ENST00000539472.1 |
SLC5A11
|
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr15_-_41120896 | 1.75 |
ENST00000299174.5
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr14_+_23352374 | 1.75 |
ENST00000267396.4
ENST00000536884.1 |
REM2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr1_-_15911510 | 1.73 |
ENST00000375826.3
|
AGMAT
|
agmatine ureohydrolase (agmatinase) |
| chr17_-_7082668 | 1.72 |
ENST00000573083.1
ENST00000574388.1 |
ASGR1
|
asialoglycoprotein receptor 1 |
| chr16_+_24857309 | 1.72 |
ENST00000565769.1
ENST00000449109.2 ENST00000424767.2 ENST00000545376.1 ENST00000569520.1 |
SLC5A11
|
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr2_+_44502630 | 1.70 |
ENST00000410056.3
ENST00000409741.1 ENST00000409229.3 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr19_-_48867291 | 1.68 |
ENST00000435956.3
|
TMEM143
|
transmembrane protein 143 |
| chr3_-_158390282 | 1.64 |
ENST00000264265.3
|
LXN
|
latexin |
| chr1_-_211848899 | 1.59 |
ENST00000366998.3
ENST00000540251.1 ENST00000366999.4 |
NEK2
|
NIMA-related kinase 2 |
| chr19_-_39303576 | 1.58 |
ENST00000594209.1
|
LGALS4
|
lectin, galactoside-binding, soluble, 4 |
| chr6_-_86099898 | 1.53 |
ENST00000455071.1
|
RP11-30P6.6
|
RP11-30P6.6 |
| chr6_-_33385854 | 1.50 |
ENST00000488478.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr16_+_19429018 | 1.48 |
ENST00000542583.2
|
TMC5
|
transmembrane channel-like 5 |
| chr11_-_45939565 | 1.47 |
ENST00000525192.1
ENST00000378750.5 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr2_+_44502597 | 1.47 |
ENST00000260649.6
ENST00000409387.1 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr13_+_50656307 | 1.44 |
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr14_-_51561784 | 1.42 |
ENST00000360392.4
|
TRIM9
|
tripartite motif containing 9 |
| chr9_+_139839711 | 1.39 |
ENST00000224181.3
|
C8G
|
complement component 8, gamma polypeptide |
| chr6_+_43028182 | 1.37 |
ENST00000394058.1
|
KLC4
|
kinesin light chain 4 |
| chr8_-_98290087 | 1.35 |
ENST00000322128.3
|
TSPYL5
|
TSPY-like 5 |
| chr16_+_21244986 | 1.32 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr19_+_39881951 | 1.32 |
ENST00000315588.5
ENST00000594368.1 ENST00000599213.2 ENST00000596297.1 |
MED29
|
mediator complex subunit 29 |
| chr11_-_118868682 | 1.30 |
ENST00000526453.1
|
RP11-110I1.12
|
RP11-110I1.12 |
| chr19_-_15235906 | 1.29 |
ENST00000600984.1
|
ILVBL
|
ilvB (bacterial acetolactate synthase)-like |
| chr9_+_139839686 | 1.28 |
ENST00000371634.2
|
C8G
|
complement component 8, gamma polypeptide |
| chr5_-_16509101 | 1.27 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr1_+_23695680 | 1.25 |
ENST00000454117.1
ENST00000335648.3 ENST00000518821.1 ENST00000437367.2 |
C1orf213
|
chromosome 1 open reading frame 213 |
| chr11_+_74870818 | 1.24 |
ENST00000525845.1
ENST00000534186.1 ENST00000428359.2 |
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1 |
| chr14_-_51562037 | 1.24 |
ENST00000338969.5
|
TRIM9
|
tripartite motif containing 9 |
| chr2_-_61389168 | 1.23 |
ENST00000607743.1
ENST00000605902.1 |
RP11-493E12.1
|
RP11-493E12.1 |
| chr6_-_33385655 | 1.22 |
ENST00000440279.3
ENST00000607266.1 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr11_-_32456891 | 1.22 |
ENST00000452863.3
|
WT1
|
Wilms tumor 1 |
| chr1_+_94884023 | 1.21 |
ENST00000315713.5
|
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr1_+_110163202 | 1.20 |
ENST00000531203.1
ENST00000256578.3 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr11_+_64073699 | 1.19 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chrX_+_65382381 | 1.16 |
ENST00000519389.1
|
HEPH
|
hephaestin |
| chr6_-_33385823 | 1.15 |
ENST00000494751.1
ENST00000374496.3 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr6_+_30130969 | 1.14 |
ENST00000376694.4
|
TRIM15
|
tripartite motif containing 15 |
| chr7_-_78400598 | 1.13 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr8_-_16859690 | 1.11 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr16_-_88752889 | 1.11 |
ENST00000332281.5
|
SNAI3
|
snail family zinc finger 3 |
| chr1_+_45274154 | 1.09 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr11_-_64660916 | 1.08 |
ENST00000413053.1
|
MIR194-2
|
microRNA 194-2 |
| chr7_+_120628731 | 1.07 |
ENST00000310396.5
|
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr2_-_62733476 | 1.07 |
ENST00000335390.5
|
TMEM17
|
transmembrane protein 17 |
| chr4_+_109541722 | 1.06 |
ENST00000394667.3
ENST00000502534.1 |
RPL34
|
ribosomal protein L34 |
| chr22_-_42526802 | 1.06 |
ENST00000359033.4
ENST00000389970.3 ENST00000360608.5 |
CYP2D6
|
cytochrome P450, family 2, subfamily D, polypeptide 6 |
| chr19_+_751122 | 1.03 |
ENST00000215582.6
|
MISP
|
mitotic spindle positioning |
| chrX_+_135388147 | 1.02 |
ENST00000394141.1
|
GPR112
|
G protein-coupled receptor 112 |
| chr1_+_241695670 | 1.02 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr15_-_101142401 | 1.01 |
ENST00000314742.8
|
LINS
|
lines homolog (Drosophila) |
| chr20_-_44485835 | 1.00 |
ENST00000457981.1
ENST00000426915.1 ENST00000217455.4 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr3_+_46919235 | 0.98 |
ENST00000449590.1
|
PTH1R
|
parathyroid hormone 1 receptor |
| chr17_-_2614927 | 0.97 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr17_-_27503770 | 0.97 |
ENST00000533112.1
|
MYO18A
|
myosin XVIIIA |
| chr8_+_22436635 | 0.95 |
ENST00000452226.1
ENST00000397760.4 ENST00000339162.7 ENST00000397761.2 |
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr17_-_47786375 | 0.95 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr2_-_27603582 | 0.95 |
ENST00000323703.6
ENST00000436006.1 |
ZNF513
|
zinc finger protein 513 |
| chr16_+_2255841 | 0.94 |
ENST00000301725.7
|
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr12_-_121973974 | 0.93 |
ENST00000538379.1
ENST00000541318.1 ENST00000541511.1 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr1_+_113217309 | 0.93 |
ENST00000544796.1
ENST00000369644.1 |
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr19_-_14201776 | 0.92 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr1_-_167059830 | 0.88 |
ENST00000367868.3
|
GPA33
|
glycoprotein A33 (transmembrane) |
| chr2_+_128293323 | 0.88 |
ENST00000389524.4
ENST00000428314.1 |
MYO7B
|
myosin VIIB |
| chr12_-_21757774 | 0.87 |
ENST00000261195.2
|
GYS2
|
glycogen synthase 2 (liver) |
| chr19_+_41949054 | 0.86 |
ENST00000378187.2
|
C19orf69
|
chromosome 19 open reading frame 69 |
| chrX_+_65382433 | 0.86 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr16_+_2255710 | 0.85 |
ENST00000397124.1
ENST00000565250.1 |
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr2_-_61389240 | 0.85 |
ENST00000606876.1
|
RP11-493E12.1
|
RP11-493E12.1 |
| chr10_-_21186144 | 0.85 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr10_+_102106829 | 0.80 |
ENST00000370355.2
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr17_+_44039704 | 0.77 |
ENST00000420682.2
ENST00000415613.2 ENST00000571987.1 ENST00000574436.1 ENST00000431008.3 |
MAPT
|
microtubule-associated protein tau |
| chr1_-_177939348 | 0.77 |
ENST00000464631.2
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr1_+_241695424 | 0.77 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr15_-_101142362 | 0.76 |
ENST00000559577.1
ENST00000561308.1 ENST00000560133.1 ENST00000560941.1 ENST00000559736.1 ENST00000560272.1 |
LINS
|
lines homolog (Drosophila) |
| chr1_+_113217073 | 0.75 |
ENST00000369645.1
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr1_-_43855479 | 0.74 |
ENST00000290663.6
ENST00000372457.4 |
MED8
|
mediator complex subunit 8 |
| chr10_+_18689637 | 0.73 |
ENST00000377315.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr5_+_1801503 | 0.73 |
ENST00000274137.5
ENST00000469176.1 |
NDUFS6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) |
| chr9_-_35685452 | 0.73 |
ENST00000607559.1
|
TPM2
|
tropomyosin 2 (beta) |
| chr1_-_41131326 | 0.72 |
ENST00000372684.3
|
RIMS3
|
regulating synaptic membrane exocytosis 3 |
| chr16_+_24857162 | 0.72 |
ENST00000347898.3
|
SLC5A11
|
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr17_+_1646130 | 0.72 |
ENST00000453066.1
ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr20_+_57875758 | 0.70 |
ENST00000395654.3
|
EDN3
|
endothelin 3 |
| chr20_+_56136136 | 0.70 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr20_+_57875658 | 0.70 |
ENST00000371025.3
|
EDN3
|
endothelin 3 |
| chr17_+_41857793 | 0.68 |
ENST00000449302.3
|
C17orf105
|
chromosome 17 open reading frame 105 |
| chr6_+_131148538 | 0.63 |
ENST00000541421.2
|
SMLR1
|
small leucine-rich protein 1 |
| chr1_+_113217043 | 0.61 |
ENST00000413052.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr19_-_42758040 | 0.59 |
ENST00000593944.1
|
ERF
|
Ets2 repressor factor |
| chr3_+_108541608 | 0.57 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr19_+_4639514 | 0.54 |
ENST00000327473.4
|
TNFAIP8L1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr6_+_30881982 | 0.54 |
ENST00000321897.5
ENST00000416670.2 ENST00000542001.1 ENST00000428017.1 |
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr16_+_66600294 | 0.53 |
ENST00000535705.1
ENST00000332695.7 ENST00000336328.6 ENST00000528324.1 ENST00000531885.1 ENST00000529506.1 ENST00000457188.2 ENST00000533666.1 ENST00000533953.1 ENST00000379500.2 ENST00000328020.6 |
CMTM1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chr10_+_70320413 | 0.53 |
ENST00000373644.4
|
TET1
|
tet methylcytosine dioxygenase 1 |
| chr7_-_75452673 | 0.53 |
ENST00000416943.1
|
CCL24
|
chemokine (C-C motif) ligand 24 |
| chr1_-_43855444 | 0.51 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr14_+_100485712 | 0.51 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr16_+_29690358 | 0.51 |
ENST00000395384.4
ENST00000562473.1 |
QPRT
|
quinolinate phosphoribosyltransferase |
| chr11_-_71781096 | 0.51 |
ENST00000535087.1
ENST00000535838.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr5_+_136070614 | 0.50 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr1_+_110163682 | 0.50 |
ENST00000358729.4
|
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr1_+_110163709 | 0.50 |
ENST00000369840.2
ENST00000527846.1 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr3_+_108541545 | 0.49 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr3_-_125775629 | 0.48 |
ENST00000383598.2
|
SLC41A3
|
solute carrier family 41, member 3 |
| chr22_+_30163340 | 0.48 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr22_+_27068704 | 0.48 |
ENST00000444388.1
ENST00000450963.1 ENST00000449017.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr7_+_33765593 | 0.46 |
ENST00000311067.3
|
RP11-89N17.1
|
HCG1643653; Uncharacterized protein |
| chr1_+_48688357 | 0.44 |
ENST00000533824.1
ENST00000438567.2 ENST00000236495.5 ENST00000420136.2 |
SLC5A9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr5_-_173043591 | 0.44 |
ENST00000285908.5
ENST00000480951.1 ENST00000311086.4 |
BOD1
|
biorientation of chromosomes in cell division 1 |
| chr1_+_100435535 | 0.43 |
ENST00000427993.2
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr17_-_28618867 | 0.43 |
ENST00000394819.3
ENST00000577623.1 |
BLMH
|
bleomycin hydrolase |
| chr7_-_78400364 | 0.43 |
ENST00000535697.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr1_+_145727681 | 0.42 |
ENST00000417171.1
ENST00000451928.2 |
PDZK1
|
PDZ domain containing 1 |
| chr1_-_41131106 | 0.42 |
ENST00000372683.1
|
RIMS3
|
regulating synaptic membrane exocytosis 3 |
| chr8_-_79717163 | 0.40 |
ENST00000520269.1
|
IL7
|
interleukin 7 |
| chr20_-_36661826 | 0.40 |
ENST00000373448.2
ENST00000373447.3 |
TTI1
|
TELO2 interacting protein 1 |
| chr6_+_30882108 | 0.40 |
ENST00000541562.1
ENST00000421263.1 |
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr15_-_100273544 | 0.39 |
ENST00000409796.1
ENST00000545021.1 ENST00000344791.2 ENST00000332728.4 ENST00000450512.1 |
LYSMD4
|
LysM, putative peptidoglycan-binding, domain containing 4 |
| chr4_+_109541772 | 0.39 |
ENST00000506397.1
ENST00000394668.2 |
RPL34
|
ribosomal protein L34 |
| chr11_+_118868830 | 0.37 |
ENST00000334418.1
|
CCDC84
|
coiled-coil domain containing 84 |
| chr17_+_27369918 | 0.37 |
ENST00000323372.4
|
PIPOX
|
pipecolic acid oxidase |
Network of associatons between targets according to the STRING database.
First level regulatory network of HNF4A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 24.5 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 2.0 | 6.0 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 1.6 | 4.9 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 1.6 | 3.2 | GO:0097187 | dentinogenesis(GO:0097187) |
| 1.4 | 10.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 1.1 | 3.3 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 1.1 | 9.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 1.0 | 3.1 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 1.0 | 3.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 1.0 | 4.8 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.8 | 9.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.7 | 3.6 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.7 | 2.1 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.7 | 2.7 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.6 | 4.2 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.6 | 1.8 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.6 | 2.3 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.6 | 2.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.6 | 3.9 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.5 | 2.6 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.5 | 2.0 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.4 | 3.2 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.4 | 3.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.3 | 1.3 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.3 | 14.0 | GO:0036499 | PERK-mediated unfolded protein response(GO:0036499) |
| 0.3 | 9.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.3 | 1.0 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.3 | 3.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.3 | 2.2 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.3 | 1.2 | GO:0072302 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.3 | 3.0 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.3 | 9.2 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.3 | 2.5 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.3 | 1.9 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.3 | 0.8 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.3 | 3.0 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.2 | 1.7 | GO:0008295 | spermidine biosynthetic process(GO:0008295) primary amino compound biosynthetic process(GO:1901162) |
| 0.2 | 1.0 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.2 | 2.2 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.2 | 2.7 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 1.0 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.2 | 0.7 | GO:1904640 | response to methionine(GO:1904640) |
| 0.2 | 1.6 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.2 | 4.3 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.2 | 4.2 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.2 | 2.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 0.8 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.2 | 0.9 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.2 | 1.6 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.2 | 0.9 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.2 | 0.9 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 3.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 2.8 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 3.7 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 2.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 6.0 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 1.1 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 | 0.5 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 0.4 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 0.7 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 0.7 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 0.4 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 4.9 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 1.6 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 3.1 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.2 | GO:0060621 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.1 | 0.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.7 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 1.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 2.0 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.5 | GO:0001826 | inner cell mass cell differentiation(GO:0001826) |
| 0.1 | 0.4 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.1 | 0.9 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.0 | 0.1 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.0 | 0.5 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 3.2 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 4.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 1.0 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 1.8 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 3.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 4.5 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.7 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 1.3 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 1.8 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.1 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 1.3 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 1.2 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 1.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.9 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.4 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.3 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.2 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 9.3 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.0 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.0 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.2 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 3.9 | GO:0010038 | response to metal ion(GO:0010038) |
| 0.0 | 0.2 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 2.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 3.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.2 | 20.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 2.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.6 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 4.9 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.8 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 0.3 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 3.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.5 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 10.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 5.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 1.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 3.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 2.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 17.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.5 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 8.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 7.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 3.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 2.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 10.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.5 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 3.0 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 2.0 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 14.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.0 | 3.0 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 1.0 | 3.9 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.8 | 3.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.7 | 3.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.7 | 2.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.6 | 6.0 | GO:0005497 | androgen binding(GO:0005497) |
| 0.6 | 4.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.5 | 4.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.5 | 4.6 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.5 | 9.1 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.5 | 4.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.4 | 2.8 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.4 | 2.0 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.4 | 3.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.3 | 3.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.3 | 2.0 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.3 | 2.3 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.3 | 4.8 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.3 | 3.0 | GO:0030249 | cyclase regulator activity(GO:0010851) guanylate cyclase regulator activity(GO:0030249) |
| 0.3 | 2.5 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.3 | 5.0 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.3 | 1.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 1.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.2 | 1.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.2 | 6.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 1.0 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.2 | 9.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 0.7 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.2 | 1.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 2.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 5.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.2 | 1.0 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.2 | 0.8 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.2 | 7.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 1.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.2 | 2.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.9 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.2 | 3.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 2.7 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 3.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.5 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.5 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 1.8 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 4.9 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 1.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.5 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 21.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 1.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 1.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.7 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.8 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 20.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 4.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.3 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.1 | 3.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.9 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.4 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 0.5 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.2 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 3.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.4 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 2.6 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 4.9 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.0 | 0.9 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 1.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 1.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 1.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 3.2 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 2.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 9.5 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 11.7 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.7 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.0 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 38.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 6.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 6.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 3.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 4.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 4.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 3.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 2.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 9.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 3.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.2 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 34.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.5 | 14.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 5.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 8.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 9.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 3.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 3.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 4.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 5.2 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 2.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 2.7 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 2.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 4.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 4.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 2.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 0.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 2.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 3.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 2.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.6 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 1.8 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.7 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 4.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 3.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 3.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 6.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 2.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 3.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.9 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |