Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
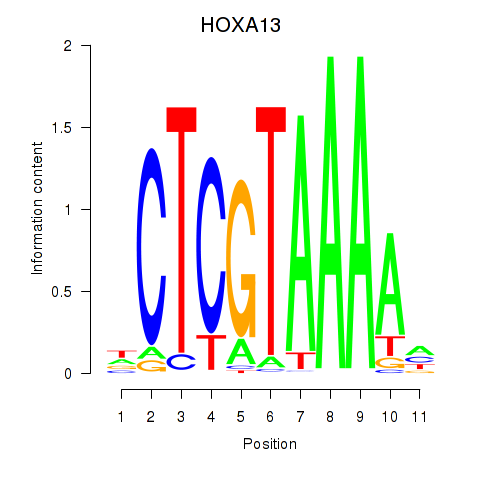
Results for HOXA13
Z-value: 0.47
Transcription factors associated with HOXA13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA13
|
ENSG00000106031.6 | homeobox A13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA13 | hg19_v2_chr7_-_27239703_27239725 | 0.80 | 2.4e-05 | Click! |
Activity profile of HOXA13 motif
Sorted Z-values of HOXA13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_37554955 | 1.40 |
ENST00000217429.4
|
FAM83D
|
family with sequence similarity 83, member D |
| chr2_-_225811747 | 1.07 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr4_-_40516560 | 0.93 |
ENST00000513473.1
|
RBM47
|
RNA binding motif protein 47 |
| chr3_+_160117062 | 0.77 |
ENST00000497311.1
|
SMC4
|
structural maintenance of chromosomes 4 |
| chr2_+_37571717 | 0.69 |
ENST00000338415.3
ENST00000404976.1 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr8_-_95274536 | 0.63 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr2_-_25100893 | 0.61 |
ENST00000433852.1
|
ADCY3
|
adenylate cyclase 3 |
| chrX_-_20236970 | 0.60 |
ENST00000379548.4
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr12_-_116714564 | 0.60 |
ENST00000548743.1
|
MED13L
|
mediator complex subunit 13-like |
| chr5_+_169010638 | 0.57 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr13_+_34392200 | 0.56 |
ENST00000434425.1
|
RFC3
|
replication factor C (activator 1) 3, 38kDa |
| chr9_+_67977438 | 0.55 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr1_+_199996733 | 0.55 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr4_+_106631966 | 0.55 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr4_+_48485341 | 0.55 |
ENST00000273861.4
|
SLC10A4
|
solute carrier family 10, member 4 |
| chr17_+_17685422 | 0.54 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr12_+_20963632 | 0.52 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr15_+_85523671 | 0.52 |
ENST00000310298.4
ENST00000557957.1 |
PDE8A
|
phosphodiesterase 8A |
| chr16_+_19619083 | 0.52 |
ENST00000538552.1
|
C16orf62
|
chromosome 16 open reading frame 62 |
| chrX_-_19688475 | 0.51 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr2_+_37571845 | 0.50 |
ENST00000537448.1
|
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr17_+_57233087 | 0.50 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr5_-_58571935 | 0.49 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr1_+_222886694 | 0.47 |
ENST00000426638.1
ENST00000537020.1 ENST00000539697.1 |
BROX
|
BRO1 domain and CAAX motif containing |
| chr17_-_46806540 | 0.47 |
ENST00000290295.7
|
HOXB13
|
homeobox B13 |
| chr5_+_138210919 | 0.47 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr11_+_111896090 | 0.44 |
ENST00000393051.1
|
DLAT
|
dihydrolipoamide S-acetyltransferase |
| chr13_+_34392185 | 0.44 |
ENST00000380071.3
|
RFC3
|
replication factor C (activator 1) 3, 38kDa |
| chr20_-_25320367 | 0.44 |
ENST00000450393.1
ENST00000491682.1 |
ABHD12
|
abhydrolase domain containing 12 |
| chr12_-_47473425 | 0.44 |
ENST00000550413.1
|
AMIGO2
|
adhesion molecule with Ig-like domain 2 |
| chr17_-_76824975 | 0.43 |
ENST00000586066.2
|
USP36
|
ubiquitin specific peptidase 36 |
| chr16_+_53483983 | 0.43 |
ENST00000544545.1
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr14_-_80678512 | 0.43 |
ENST00000553968.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr3_-_123339343 | 0.42 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr2_+_109223595 | 0.42 |
ENST00000410093.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr3_+_113667354 | 0.42 |
ENST00000491556.1
|
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr9_+_137979506 | 0.41 |
ENST00000539529.1
ENST00000392991.4 ENST00000371793.3 |
OLFM1
|
olfactomedin 1 |
| chr7_-_27196267 | 0.41 |
ENST00000242159.3
|
HOXA7
|
homeobox A7 |
| chr3_-_8811288 | 0.41 |
ENST00000316793.3
ENST00000431493.1 |
OXTR
|
oxytocin receptor |
| chr16_-_28192360 | 0.41 |
ENST00000570033.1
|
XPO6
|
exportin 6 |
| chr19_+_36545781 | 0.41 |
ENST00000388999.3
|
WDR62
|
WD repeat domain 62 |
| chr11_+_111896320 | 0.41 |
ENST00000531306.1
ENST00000537636.1 |
DLAT
|
dihydrolipoamide S-acetyltransferase |
| chr2_+_7073174 | 0.40 |
ENST00000416587.1
|
RNF144A
|
ring finger protein 144A |
| chr19_+_36545833 | 0.40 |
ENST00000401500.2
ENST00000270301.7 ENST00000427823.2 |
WDR62
|
WD repeat domain 62 |
| chr15_-_88799661 | 0.39 |
ENST00000360948.2
ENST00000357724.2 ENST00000355254.2 ENST00000317501.3 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr17_-_46688334 | 0.39 |
ENST00000239165.7
|
HOXB7
|
homeobox B7 |
| chr11_+_61891445 | 0.39 |
ENST00000394818.3
ENST00000533896.1 ENST00000278849.4 |
INCENP
|
inner centromere protein antigens 135/155kDa |
| chr5_-_146781153 | 0.39 |
ENST00000520473.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr15_+_59908633 | 0.38 |
ENST00000559626.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr17_+_55183261 | 0.37 |
ENST00000576295.1
|
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr16_-_18908196 | 0.36 |
ENST00000565324.1
ENST00000561947.1 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr2_+_174219548 | 0.36 |
ENST00000347703.3
ENST00000392567.2 ENST00000306721.3 ENST00000410101.3 ENST00000410019.3 |
CDCA7
|
cell division cycle associated 7 |
| chr1_+_173793777 | 0.36 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr12_+_97300995 | 0.36 |
ENST00000266742.4
ENST00000429527.2 ENST00000554226.1 ENST00000557478.1 ENST00000557092.1 ENST00000411739.2 ENST00000553609.1 |
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr4_+_141178440 | 0.35 |
ENST00000394205.3
|
SCOC
|
short coiled-coil protein |
| chr11_-_115127611 | 0.35 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chr17_-_57158523 | 0.35 |
ENST00000581468.1
|
TRIM37
|
tripartite motif containing 37 |
| chr17_-_40170506 | 0.35 |
ENST00000589773.1
ENST00000590348.1 |
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr3_-_123339418 | 0.35 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr11_+_34999328 | 0.34 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr12_+_47473369 | 0.33 |
ENST00000546455.1
|
PCED1B
|
PC-esterase domain containing 1B |
| chrX_-_10645724 | 0.33 |
ENST00000413894.1
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr14_+_56078695 | 0.33 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr19_+_34887220 | 0.33 |
ENST00000592740.1
|
RP11-618P17.4
|
Uncharacterized protein |
| chr12_+_45609862 | 0.32 |
ENST00000423947.3
ENST00000435642.1 |
ANO6
|
anoctamin 6 |
| chr2_-_133429091 | 0.32 |
ENST00000345008.6
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chrY_+_2709527 | 0.32 |
ENST00000250784.8
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1 |
| chr12_+_45609797 | 0.32 |
ENST00000425752.2
|
ANO6
|
anoctamin 6 |
| chr3_+_99979828 | 0.31 |
ENST00000485687.1
ENST00000344949.5 ENST00000394144.4 |
TBC1D23
|
TBC1 domain family, member 23 |
| chr12_+_20963647 | 0.30 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr2_+_32390925 | 0.30 |
ENST00000440718.1
ENST00000379343.2 ENST00000282587.5 ENST00000435660.1 ENST00000538303.1 ENST00000357055.3 ENST00000406369.1 |
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr1_+_199996702 | 0.30 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr1_+_100818009 | 0.29 |
ENST00000370125.2
ENST00000361544.6 ENST00000370124.3 |
CDC14A
|
cell division cycle 14A |
| chr1_-_207224307 | 0.29 |
ENST00000315927.4
|
YOD1
|
YOD1 deubiquitinase |
| chr22_+_45714301 | 0.29 |
ENST00000427777.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr3_-_118753716 | 0.29 |
ENST00000393775.2
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chrX_+_9431324 | 0.29 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr19_+_35773242 | 0.28 |
ENST00000222304.3
|
HAMP
|
hepcidin antimicrobial peptide |
| chr3_+_57882024 | 0.28 |
ENST00000494088.1
|
SLMAP
|
sarcolemma associated protein |
| chr8_-_110346614 | 0.28 |
ENST00000239690.4
|
NUDCD1
|
NudC domain containing 1 |
| chr2_+_182756615 | 0.27 |
ENST00000431877.2
ENST00000320370.7 |
SSFA2
|
sperm specific antigen 2 |
| chr10_-_30638090 | 0.27 |
ENST00000421701.1
ENST00000263063.4 |
MTPAP
|
mitochondrial poly(A) polymerase |
| chr3_+_99986036 | 0.27 |
ENST00000471098.1
|
TBC1D23
|
TBC1 domain family, member 23 |
| chr5_-_78808617 | 0.27 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chr3_-_118753626 | 0.26 |
ENST00000489689.1
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr17_-_17480779 | 0.26 |
ENST00000395782.1
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr1_-_150208291 | 0.26 |
ENST00000533654.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr1_-_230991747 | 0.26 |
ENST00000523410.1
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr10_-_113943447 | 0.26 |
ENST00000369425.1
ENST00000348367.4 ENST00000423155.1 |
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr8_-_29939933 | 0.25 |
ENST00000522794.1
|
TMEM66
|
transmembrane protein 66 |
| chr15_+_77713299 | 0.25 |
ENST00000559099.1
|
HMG20A
|
high mobility group 20A |
| chr1_-_150208498 | 0.25 |
ENST00000314136.8
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr22_+_18632666 | 0.25 |
ENST00000215794.7
|
USP18
|
ubiquitin specific peptidase 18 |
| chr5_-_40755987 | 0.25 |
ENST00000337702.4
|
TTC33
|
tetratricopeptide repeat domain 33 |
| chr7_-_73256838 | 0.25 |
ENST00000297873.4
|
WBSCR27
|
Williams Beuren syndrome chromosome region 27 |
| chr14_-_20774092 | 0.25 |
ENST00000423949.2
ENST00000553828.1 ENST00000258821.3 |
TTC5
|
tetratricopeptide repeat domain 5 |
| chr15_+_71228826 | 0.24 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr1_-_150208412 | 0.24 |
ENST00000532744.1
ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr5_+_133861790 | 0.23 |
ENST00000395003.1
|
PHF15
|
jade family PHD finger 2 |
| chr1_+_84609944 | 0.23 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr2_+_33359473 | 0.23 |
ENST00000432635.1
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr3_-_149093499 | 0.23 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr20_+_19870167 | 0.23 |
ENST00000440354.2
|
RIN2
|
Ras and Rab interactor 2 |
| chr15_+_52155001 | 0.22 |
ENST00000544199.1
|
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr17_-_40021656 | 0.22 |
ENST00000319121.3
|
KLHL11
|
kelch-like family member 11 |
| chr7_-_69062391 | 0.22 |
ENST00000436600.2
|
RP5-942I16.1
|
RP5-942I16.1 |
| chr15_-_55881135 | 0.22 |
ENST00000302000.6
|
PYGO1
|
pygopus family PHD finger 1 |
| chr2_-_74007095 | 0.22 |
ENST00000452812.1
|
DUSP11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr4_+_122722466 | 0.22 |
ENST00000243498.5
ENST00000379663.3 ENST00000509800.1 |
EXOSC9
|
exosome component 9 |
| chr15_-_99789736 | 0.21 |
ENST00000560235.1
ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr6_+_121756809 | 0.20 |
ENST00000282561.3
|
GJA1
|
gap junction protein, alpha 1, 43kDa |
| chr9_-_116172617 | 0.20 |
ENST00000374169.3
|
POLE3
|
polymerase (DNA directed), epsilon 3, accessory subunit |
| chr19_-_14530112 | 0.20 |
ENST00000592632.1
ENST00000589675.1 |
DDX39A
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39A |
| chr1_-_150208320 | 0.20 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr13_+_78315466 | 0.20 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chrX_+_123097014 | 0.20 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr5_+_140180635 | 0.20 |
ENST00000522353.2
ENST00000532566.2 |
PCDHA3
|
protocadherin alpha 3 |
| chr20_+_55204351 | 0.19 |
ENST00000201031.2
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chrX_-_135962923 | 0.19 |
ENST00000565438.1
|
RBMX
|
RNA binding motif protein, X-linked |
| chr16_-_66764119 | 0.19 |
ENST00000569320.1
|
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr9_-_139137648 | 0.19 |
ENST00000358701.5
|
QSOX2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr6_+_134273300 | 0.19 |
ENST00000416965.1
|
TBPL1
|
TBP-like 1 |
| chrX_-_135962876 | 0.19 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chr3_+_101498269 | 0.19 |
ENST00000491511.2
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr4_-_17513604 | 0.19 |
ENST00000505710.1
|
QDPR
|
quinoid dihydropteridine reductase |
| chr2_+_33359687 | 0.18 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr1_-_150208363 | 0.18 |
ENST00000436748.2
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr17_-_15496722 | 0.18 |
ENST00000472534.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr10_-_13544945 | 0.18 |
ENST00000378605.3
ENST00000341083.3 |
BEND7
|
BEN domain containing 7 |
| chr10_+_90750493 | 0.18 |
ENST00000357339.2
ENST00000355279.2 |
FAS
|
Fas cell surface death receptor |
| chr5_+_95997885 | 0.18 |
ENST00000511097.2
|
CAST
|
calpastatin |
| chr9_-_21305312 | 0.17 |
ENST00000259555.4
|
IFNA5
|
interferon, alpha 5 |
| chr19_-_6279932 | 0.17 |
ENST00000252674.7
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 |
| chr10_-_52383644 | 0.17 |
ENST00000361781.2
|
SGMS1
|
sphingomyelin synthase 1 |
| chr14_-_71067360 | 0.17 |
ENST00000554963.1
ENST00000430055.2 ENST00000440435.2 ENST00000256379.5 |
MED6
|
mediator complex subunit 6 |
| chr9_-_98269481 | 0.17 |
ENST00000418258.1
ENST00000553011.1 ENST00000551845.1 |
PTCH1
|
patched 1 |
| chr16_+_66914264 | 0.17 |
ENST00000311765.2
ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr5_+_68860949 | 0.16 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr3_+_111578027 | 0.16 |
ENST00000431670.2
ENST00000412622.1 |
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr2_+_33359646 | 0.16 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr3_-_118753792 | 0.16 |
ENST00000480431.1
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr1_+_153631130 | 0.16 |
ENST00000368685.5
|
SNAPIN
|
SNAP-associated protein |
| chr4_-_128887069 | 0.16 |
ENST00000541133.1
ENST00000296468.3 ENST00000513559.1 |
MFSD8
|
major facilitator superfamily domain containing 8 |
| chr5_-_79287060 | 0.16 |
ENST00000512560.1
ENST00000509852.1 ENST00000512528.1 |
MTX3
|
metaxin 3 |
| chrX_+_100878079 | 0.16 |
ENST00000471229.2
|
ARMCX3
|
armadillo repeat containing, X-linked 3 |
| chr17_+_77030267 | 0.15 |
ENST00000581774.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr16_-_1538765 | 0.15 |
ENST00000447419.2
ENST00000440447.2 |
PTX4
|
pentraxin 4, long |
| chr2_+_64681103 | 0.15 |
ENST00000464281.1
|
LGALSL
|
lectin, galactoside-binding-like |
| chr12_-_4647606 | 0.15 |
ENST00000261250.3
ENST00000541014.1 ENST00000545746.1 ENST00000542080.1 |
C12orf4
|
chromosome 12 open reading frame 4 |
| chr17_-_39538550 | 0.15 |
ENST00000394001.1
|
KRT34
|
keratin 34 |
| chr14_+_50234309 | 0.15 |
ENST00000298307.5
|
KLHDC2
|
kelch domain containing 2 |
| chr10_+_92631709 | 0.15 |
ENST00000413330.1
ENST00000277882.3 |
RPP30
|
ribonuclease P/MRP 30kDa subunit |
| chr7_-_151511911 | 0.15 |
ENST00000392801.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr3_+_129159039 | 0.14 |
ENST00000507564.1
ENST00000431818.2 ENST00000504021.1 ENST00000349441.2 ENST00000348417.2 ENST00000440957.2 |
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr19_+_49497121 | 0.14 |
ENST00000413176.2
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr1_+_179050512 | 0.14 |
ENST00000367627.3
|
TOR3A
|
torsin family 3, member A |
| chrX_-_38186775 | 0.14 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr2_-_169104651 | 0.14 |
ENST00000355999.4
|
STK39
|
serine threonine kinase 39 |
| chr14_+_74353320 | 0.14 |
ENST00000540593.1
ENST00000555730.1 |
ZNF410
|
zinc finger protein 410 |
| chr8_+_26150628 | 0.14 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr3_-_160117035 | 0.14 |
ENST00000489004.1
ENST00000496589.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr13_+_78315528 | 0.14 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr1_-_155948890 | 0.14 |
ENST00000471589.1
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr7_+_129015671 | 0.13 |
ENST00000466993.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr7_-_23145288 | 0.13 |
ENST00000419813.1
|
KLHL7-AS1
|
KLHL7 antisense RNA 1 (head to head) |
| chr14_-_92414294 | 0.13 |
ENST00000554468.1
|
FBLN5
|
fibulin 5 |
| chr17_+_33895090 | 0.13 |
ENST00000592381.1
|
RP11-1094M14.11
|
RP11-1094M14.11 |
| chr6_+_4706368 | 0.13 |
ENST00000328908.5
|
CDYL
|
chromodomain protein, Y-like |
| chr19_+_49496782 | 0.13 |
ENST00000601968.1
ENST00000596837.1 |
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr17_+_16284604 | 0.13 |
ENST00000395839.1
ENST00000395837.1 |
UBB
|
ubiquitin B |
| chr1_+_171750776 | 0.13 |
ENST00000458517.1
ENST00000362019.3 ENST00000367737.5 ENST00000361735.3 |
METTL13
|
methyltransferase like 13 |
| chr9_-_130889990 | 0.12 |
ENST00000449878.1
|
PTGES2
|
prostaglandin E synthase 2 |
| chr13_+_78315295 | 0.12 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr3_+_57882061 | 0.12 |
ENST00000461354.1
ENST00000466255.1 |
SLMAP
|
sarcolemma associated protein |
| chr19_-_14529847 | 0.12 |
ENST00000590239.1
ENST00000590696.1 ENST00000591275.1 ENST00000586993.1 |
DDX39A
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39A |
| chr4_+_128702969 | 0.12 |
ENST00000508776.1
ENST00000439123.2 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chr14_-_92414055 | 0.12 |
ENST00000342058.4
|
FBLN5
|
fibulin 5 |
| chr13_+_78315348 | 0.11 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr19_+_54369434 | 0.11 |
ENST00000421337.1
|
MYADM
|
myeloid-associated differentiation marker |
| chr3_-_170587974 | 0.11 |
ENST00000463836.1
|
RPL22L1
|
ribosomal protein L22-like 1 |
| chr3_-_170587815 | 0.11 |
ENST00000466674.1
|
RPL22L1
|
ribosomal protein L22-like 1 |
| chr1_-_217804377 | 0.11 |
ENST00000366935.3
ENST00000366934.3 |
GPATCH2
|
G patch domain containing 2 |
| chr2_+_26256938 | 0.11 |
ENST00000264710.4
|
RAB10
|
RAB10, member RAS oncogene family |
| chr12_+_45609893 | 0.11 |
ENST00000320560.8
|
ANO6
|
anoctamin 6 |
| chrX_+_117480036 | 0.11 |
ENST00000371822.5
ENST00000254029.3 ENST00000371825.3 |
WDR44
|
WD repeat domain 44 |
| chr5_-_75008244 | 0.11 |
ENST00000510798.1
ENST00000446329.2 |
POC5
|
POC5 centriolar protein |
| chr19_+_36426452 | 0.11 |
ENST00000588831.1
|
LRFN3
|
leucine rich repeat and fibronectin type III domain containing 3 |
| chr12_-_53074182 | 0.11 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr8_-_63998590 | 0.11 |
ENST00000260116.4
|
TTPA
|
tocopherol (alpha) transfer protein |
| chr9_-_98269699 | 0.11 |
ENST00000429896.2
|
PTCH1
|
patched 1 |
| chr9_-_2844058 | 0.11 |
ENST00000397885.2
|
KIAA0020
|
KIAA0020 |
| chr20_+_37590942 | 0.10 |
ENST00000373325.2
ENST00000252011.3 ENST00000373323.4 |
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr8_+_94710789 | 0.10 |
ENST00000523475.1
|
FAM92A1
|
family with sequence similarity 92, member A1 |
| chr3_-_160116995 | 0.10 |
ENST00000465537.1
ENST00000486856.1 ENST00000468218.1 ENST00000478370.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr11_+_101983176 | 0.10 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr14_+_24540154 | 0.10 |
ENST00000559778.1
ENST00000560761.1 ENST00000557889.1 |
CPNE6
|
copine VI (neuronal) |
| chr3_-_123680246 | 0.10 |
ENST00000488653.2
|
CCDC14
|
coiled-coil domain containing 14 |
| chr17_+_78234625 | 0.10 |
ENST00000508628.2
ENST00000582970.1 ENST00000456466.1 ENST00000319921.4 |
RNF213
|
ring finger protein 213 |
| chr16_-_11876408 | 0.10 |
ENST00000396516.2
|
ZC3H7A
|
zinc finger CCCH-type containing 7A |
| chr6_-_3912207 | 0.09 |
ENST00000566733.1
|
RP1-140K8.5
|
RP1-140K8.5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA13
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.2 | 0.6 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.8 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.2 | 0.6 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.1 | 0.4 | GO:0034059 | response to anoxia(GO:0034059) positive regulation of penile erection(GO:0060406) |
| 0.1 | 0.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 1.0 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.8 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.5 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.4 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.4 | GO:0048691 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.1 | 0.4 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.1 | 0.3 | GO:1904479 | cellular response to bile acid(GO:1903413) negative regulation of intestinal absorption(GO:1904479) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.5 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.4 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.3 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.1 | 0.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.3 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 1.4 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.4 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 0.3 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.8 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 1.1 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.4 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.3 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.2 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 0.2 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.1 | 0.6 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.5 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.2 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 | 0.4 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.2 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.4 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.4 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.6 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 2.3 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.2 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.5 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.0 | 0.2 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0090212 | vitamin E metabolic process(GO:0042360) regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.0 | 0.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.8 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.4 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.2 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 1.1 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.0 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.1 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.5 | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.4 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.5 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.3 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.1 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 1.0 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 1.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.4 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.8 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.3 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.4 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 1.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.6 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.5 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.6 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.5 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.0 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.3 | 0.9 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.1 | 0.4 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.1 | 1.0 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.6 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.3 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.1 | 0.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.3 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.1 | 0.3 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 0.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.3 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.3 | GO:0032551 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.1 | 0.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.4 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.3 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.2 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.1 | 0.4 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.4 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.3 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.4 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.4 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.4 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.5 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.1 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.0 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |