Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
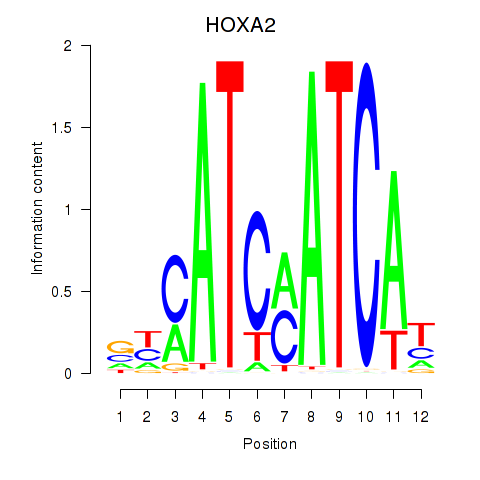
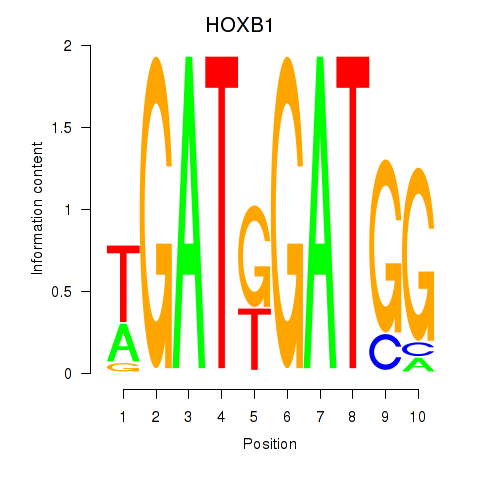
Results for HOXA2_HOXB1
Z-value: 0.59
Transcription factors associated with HOXA2_HOXB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA2
|
ENSG00000105996.5 | homeobox A2 |
|
HOXB1
|
ENSG00000120094.6 | homeobox B1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA2 | hg19_v2_chr7_-_27142290_27142430 | -0.78 | 4.7e-05 | Click! |
| HOXB1 | hg19_v2_chr17_-_46608272_46608385 | 0.62 | 3.8e-03 | Click! |
Activity profile of HOXA2_HOXB1 motif
Sorted Z-values of HOXA2_HOXB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_57365150 | 2.60 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr4_-_90756769 | 2.55 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr4_-_90757364 | 2.54 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr3_+_189507460 | 2.19 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr14_-_81893734 | 2.07 |
ENST00000555447.1
|
STON2
|
stonin 2 |
| chr3_+_189507523 | 2.02 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr10_-_105615164 | 1.85 |
ENST00000355946.2
ENST00000369774.4 |
SH3PXD2A
|
SH3 and PX domains 2A |
| chr2_+_12857043 | 1.68 |
ENST00000381465.2
|
TRIB2
|
tribbles pseudokinase 2 |
| chr3_+_189507432 | 1.63 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chr4_-_102267953 | 1.58 |
ENST00000523694.2
ENST00000507176.1 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr2_+_12857015 | 1.50 |
ENST00000155926.4
|
TRIB2
|
tribbles pseudokinase 2 |
| chr5_+_167181917 | 1.47 |
ENST00000519204.1
|
TENM2
|
teneurin transmembrane protein 2 |
| chr12_-_28124903 | 1.41 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr15_-_93616892 | 1.34 |
ENST00000556658.1
ENST00000538818.1 ENST00000425933.2 |
RGMA
|
repulsive guidance molecule family member a |
| chr15_-_93616340 | 1.34 |
ENST00000557420.1
ENST00000542321.2 |
RGMA
|
repulsive guidance molecule family member a |
| chr12_-_28125638 | 1.33 |
ENST00000545234.1
|
PTHLH
|
parathyroid hormone-like hormone |
| chrX_-_30993201 | 1.23 |
ENST00000288422.2
ENST00000378932.2 |
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr1_-_8763278 | 1.16 |
ENST00000468247.1
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr17_+_7758374 | 1.12 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr5_-_125930929 | 1.11 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr1_+_68150744 | 1.09 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr19_-_51512804 | 1.02 |
ENST00000594211.1
ENST00000376832.4 |
KLK9
|
kallikrein-related peptidase 9 |
| chr7_-_27170352 | 1.00 |
ENST00000428284.2
ENST00000360046.5 |
HOXA4
|
homeobox A4 |
| chr9_+_132099158 | 0.97 |
ENST00000444125.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr5_-_125930877 | 0.97 |
ENST00000510111.2
ENST00000509270.1 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr14_-_24733444 | 0.95 |
ENST00000560478.1
ENST00000560443.1 |
TGM1
|
transglutaminase 1 |
| chrX_+_99899180 | 0.95 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr11_-_111782696 | 0.90 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr11_-_2162162 | 0.89 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr2_-_45236540 | 0.84 |
ENST00000303077.6
|
SIX2
|
SIX homeobox 2 |
| chr20_+_52105495 | 0.80 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr17_+_48609903 | 0.78 |
ENST00000268933.3
|
EPN3
|
epsin 3 |
| chr1_-_67390474 | 0.74 |
ENST00000371023.3
ENST00000371022.3 ENST00000371026.3 ENST00000431318.1 |
WDR78
|
WD repeat domain 78 |
| chr9_-_73029540 | 0.73 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr8_-_128231299 | 0.73 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr14_-_21516590 | 0.68 |
ENST00000555026.1
|
NDRG2
|
NDRG family member 2 |
| chr5_+_145826867 | 0.68 |
ENST00000296702.5
ENST00000394421.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr5_+_154181816 | 0.66 |
ENST00000518677.1
|
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr19_+_37997812 | 0.64 |
ENST00000542455.1
ENST00000587143.1 |
ZNF793
|
zinc finger protein 793 |
| chr19_+_13051206 | 0.60 |
ENST00000586760.1
|
CALR
|
calreticulin |
| chr10_+_102891048 | 0.58 |
ENST00000467928.2
|
TLX1
|
T-cell leukemia homeobox 1 |
| chr21_-_43430440 | 0.58 |
ENST00000398505.3
ENST00000310826.5 ENST00000449949.1 ENST00000398499.1 ENST00000398497.2 ENST00000398511.3 |
ZBTB21
|
zinc finger and BTB domain containing 21 |
| chr1_-_109584608 | 0.56 |
ENST00000400794.3
ENST00000528747.1 ENST00000369962.3 ENST00000361054.3 |
WDR47
|
WD repeat domain 47 |
| chr9_+_124329336 | 0.56 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr17_+_36584662 | 0.55 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr10_+_24755416 | 0.52 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr11_-_111782484 | 0.52 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr1_+_178062855 | 0.49 |
ENST00000448150.3
|
RASAL2
|
RAS protein activator like 2 |
| chr1_+_36621174 | 0.48 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr11_+_68228186 | 0.46 |
ENST00000393799.2
ENST00000393800.2 ENST00000528635.1 ENST00000533127.1 ENST00000529907.1 ENST00000529344.1 ENST00000534534.1 ENST00000524845.1 ENST00000265637.4 ENST00000524904.1 ENST00000393801.3 ENST00000265636.5 ENST00000529710.1 |
PPP6R3
|
protein phosphatase 6, regulatory subunit 3 |
| chr1_-_205391178 | 0.45 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr1_-_109584768 | 0.44 |
ENST00000357672.3
|
WDR47
|
WD repeat domain 47 |
| chr9_-_26946981 | 0.43 |
ENST00000523212.1
|
PLAA
|
phospholipase A2-activating protein |
| chr7_+_20370746 | 0.43 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr8_-_101730061 | 0.42 |
ENST00000519100.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr1_+_12524965 | 0.41 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr1_-_109584716 | 0.40 |
ENST00000531337.1
ENST00000529074.1 ENST00000369965.4 |
WDR47
|
WD repeat domain 47 |
| chr12_+_10658489 | 0.37 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr7_-_143956815 | 0.36 |
ENST00000493325.1
|
OR2A7
|
olfactory receptor, family 2, subfamily A, member 7 |
| chr1_+_67390578 | 0.35 |
ENST00000371018.3
ENST00000355977.6 ENST00000357692.2 ENST00000401041.1 ENST00000371016.1 ENST00000371014.1 ENST00000371012.2 |
MIER1
|
mesoderm induction early response 1, transcriptional regulator |
| chr2_+_68961934 | 0.34 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr11_-_122929699 | 0.34 |
ENST00000526686.1
|
HSPA8
|
heat shock 70kDa protein 8 |
| chr6_+_32006042 | 0.33 |
ENST00000418967.2
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr2_+_54684327 | 0.31 |
ENST00000389980.3
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr2_+_68961905 | 0.31 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr2_+_242089833 | 0.30 |
ENST00000404405.3
ENST00000439916.1 ENST00000406106.3 ENST00000401987.1 |
PPP1R7
|
protein phosphatase 1, regulatory subunit 7 |
| chr6_-_159065741 | 0.30 |
ENST00000367085.3
ENST00000367089.3 |
DYNLT1
|
dynein, light chain, Tctex-type 1 |
| chr2_+_68962014 | 0.30 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr11_-_93583697 | 0.29 |
ENST00000409977.1
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr7_-_27135591 | 0.28 |
ENST00000343060.4
ENST00000355633.5 |
HOXA1
|
homeobox A1 |
| chr10_-_75457554 | 0.28 |
ENST00000374094.4
ENST00000443782.2 |
AGAP5
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 5 |
| chr1_-_217804377 | 0.27 |
ENST00000366935.3
ENST00000366934.3 |
GPATCH2
|
G patch domain containing 2 |
| chr6_+_32006159 | 0.25 |
ENST00000478281.1
ENST00000471671.1 ENST00000435122.2 |
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr1_+_52082751 | 0.25 |
ENST00000447887.1
ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
| chr13_-_36050819 | 0.24 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr12_+_57610562 | 0.23 |
ENST00000349394.5
|
NXPH4
|
neurexophilin 4 |
| chr16_-_49890016 | 0.22 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chr18_-_53070913 | 0.22 |
ENST00000568186.1
ENST00000564228.1 |
TCF4
|
transcription factor 4 |
| chr13_+_52586517 | 0.21 |
ENST00000523764.1
ENST00000521508.1 |
ALG11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr12_-_15038779 | 0.21 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr19_+_44645700 | 0.20 |
ENST00000592437.1
|
ZNF234
|
zinc finger protein 234 |
| chr10_-_65028817 | 0.20 |
ENST00000542921.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr5_-_179045199 | 0.19 |
ENST00000523921.1
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr9_-_26947220 | 0.19 |
ENST00000520884.1
|
PLAA
|
phospholipase A2-activating protein |
| chr17_+_37824411 | 0.19 |
ENST00000269582.2
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr9_-_15250114 | 0.19 |
ENST00000507993.1
|
TTC39B
|
tetratricopeptide repeat domain 39B |
| chr11_-_122930121 | 0.19 |
ENST00000524552.1
|
HSPA8
|
heat shock 70kDa protein 8 |
| chr13_+_97928395 | 0.18 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr7_-_86595190 | 0.18 |
ENST00000398276.2
ENST00000416314.1 ENST00000425689.1 |
KIAA1324L
|
KIAA1324-like |
| chr10_-_98945515 | 0.17 |
ENST00000371070.4
|
SLIT1
|
slit homolog 1 (Drosophila) |
| chr19_+_44645731 | 0.17 |
ENST00000426739.2
|
ZNF234
|
zinc finger protein 234 |
| chr10_-_65028938 | 0.16 |
ENST00000402544.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr10_-_98945264 | 0.16 |
ENST00000314867.5
|
SLIT1
|
slit homolog 1 (Drosophila) |
| chr9_-_101471479 | 0.16 |
ENST00000259455.2
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr15_+_81591757 | 0.15 |
ENST00000558332.1
|
IL16
|
interleukin 16 |
| chr17_+_1173853 | 0.15 |
ENST00000391429.1
|
BHLHA9
|
basic helix-loop-helix family, member a9 |
| chr11_+_133902547 | 0.15 |
ENST00000529070.1
|
RP11-713P17.3
|
RP11-713P17.3 |
| chr4_+_77356248 | 0.15 |
ENST00000296043.6
|
SHROOM3
|
shroom family member 3 |
| chr17_-_1588101 | 0.14 |
ENST00000577001.1
ENST00000572621.1 ENST00000304992.6 |
PRPF8
|
pre-mRNA processing factor 8 |
| chr10_-_98945677 | 0.14 |
ENST00000266058.4
ENST00000371041.3 |
SLIT1
|
slit homolog 1 (Drosophila) |
| chr17_-_42277203 | 0.14 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr10_-_46342921 | 0.13 |
ENST00000448048.2
|
AGAP4
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 4 |
| chr7_+_86273218 | 0.12 |
ENST00000361669.2
|
GRM3
|
glutamate receptor, metabotropic 3 |
| chr3_+_239652 | 0.12 |
ENST00000435603.1
|
CHL1
|
cell adhesion molecule L1-like |
| chr1_+_243419306 | 0.12 |
ENST00000355875.4
ENST00000391846.1 ENST00000366541.3 ENST00000343783.6 |
SDCCAG8
|
serologically defined colon cancer antigen 8 |
| chr15_+_48009541 | 0.12 |
ENST00000536845.2
ENST00000558816.1 |
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr5_+_98109322 | 0.12 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr8_-_38008783 | 0.11 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr3_-_72150076 | 0.11 |
ENST00000488545.1
ENST00000608654.1 |
LINC00877
|
long intergenic non-protein coding RNA 877 |
| chr5_-_145895753 | 0.10 |
ENST00000311104.2
|
GPR151
|
G protein-coupled receptor 151 |
| chr2_+_26624775 | 0.10 |
ENST00000288710.2
|
DRC1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr6_-_112575912 | 0.10 |
ENST00000522006.1
ENST00000230538.7 ENST00000519932.1 |
LAMA4
|
laminin, alpha 4 |
| chr6_-_112575687 | 0.10 |
ENST00000521398.1
ENST00000424408.2 ENST00000243219.3 |
LAMA4
|
laminin, alpha 4 |
| chr12_+_56414851 | 0.10 |
ENST00000547167.1
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr19_-_12997995 | 0.10 |
ENST00000264834.4
|
KLF1
|
Kruppel-like factor 1 (erythroid) |
| chr2_+_135011731 | 0.09 |
ENST00000281923.2
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr12_+_7282795 | 0.09 |
ENST00000266546.6
|
CLSTN3
|
calsyntenin 3 |
| chr9_-_26947453 | 0.09 |
ENST00000397292.3
|
PLAA
|
phospholipase A2-activating protein |
| chr19_+_52074502 | 0.09 |
ENST00000545217.1
ENST00000262259.2 ENST00000596504.1 |
ZNF175
|
zinc finger protein 175 |
| chr7_+_1272522 | 0.09 |
ENST00000316333.8
|
UNCX
|
UNC homeobox |
| chrX_+_125953746 | 0.09 |
ENST00000371125.3
|
CXorf64
|
chromosome X open reading frame 64 |
| chr3_+_150264458 | 0.08 |
ENST00000487799.1
ENST00000460851.1 |
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa |
| chr8_-_67874805 | 0.08 |
ENST00000563496.1
|
TCF24
|
transcription factor 24 |
| chr6_+_33387868 | 0.07 |
ENST00000418600.2
|
SYNGAP1
|
synaptic Ras GTPase activating protein 1 |
| chr16_-_54972581 | 0.07 |
ENST00000559802.1
ENST00000558156.1 |
CTD-3032H12.1
|
CTD-3032H12.1 |
| chr6_+_73331520 | 0.06 |
ENST00000342056.2
ENST00000355194.4 |
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr16_+_66429358 | 0.06 |
ENST00000539168.1
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr7_+_26591441 | 0.05 |
ENST00000420912.1
ENST00000457000.1 ENST00000430426.1 |
AC004947.2
|
AC004947.2 |
| chr2_-_23747214 | 0.05 |
ENST00000430988.1
|
AC011239.1
|
Uncharacterized protein |
| chr6_-_112575758 | 0.04 |
ENST00000431543.2
ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4
|
laminin, alpha 4 |
| chr12_+_57482877 | 0.04 |
ENST00000342556.6
ENST00000357680.4 |
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr3_+_191046810 | 0.04 |
ENST00000392455.3
ENST00000392456.3 |
CCDC50
|
coiled-coil domain containing 50 |
| chr20_+_30467600 | 0.04 |
ENST00000375934.4
ENST00000375922.4 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr14_+_68189190 | 0.04 |
ENST00000539142.1
|
RDH12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis) |
| chr12_+_57390745 | 0.03 |
ENST00000600202.1
|
HBCBP
|
HBcAg-binding protein; Uncharacterized protein |
| chr6_+_155334780 | 0.03 |
ENST00000538270.1
ENST00000535231.1 |
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr1_-_223536679 | 0.02 |
ENST00000608996.1
|
SUSD4
|
sushi domain containing 4 |
| chr7_-_120497178 | 0.02 |
ENST00000441017.1
ENST00000424710.1 ENST00000433758.1 |
TSPAN12
|
tetraspanin 12 |
| chr1_-_108507631 | 0.02 |
ENST00000527011.1
ENST00000370056.4 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr21_+_19617140 | 0.02 |
ENST00000299295.2
ENST00000338326.3 |
CHODL
|
chondrolectin |
| chr12_-_114843889 | 0.02 |
ENST00000405440.2
|
TBX5
|
T-box 5 |
| chr6_-_112575838 | 0.01 |
ENST00000455073.1
|
LAMA4
|
laminin, alpha 4 |
| chr2_-_154335300 | 0.01 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chrX_-_45629661 | 0.01 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr16_-_11617444 | 0.01 |
ENST00000598234.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chr12_-_86650077 | 0.01 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr20_-_45976816 | 0.01 |
ENST00000441977.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr2_-_44588679 | 0.00 |
ENST00000409411.1
|
PREPL
|
prolyl endopeptidase-like |
| chr8_-_143961236 | 0.00 |
ENST00000377675.3
ENST00000517471.1 ENST00000292427.4 |
CYP11B1
|
cytochrome P450, family 11, subfamily B, polypeptide 1 |
| chr12_-_88423164 | 0.00 |
ENST00000298699.2
ENST00000550553.1 |
C12orf50
|
chromosome 12 open reading frame 50 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA2_HOXB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.8 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 1.1 | 3.2 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 1.0 | 5.1 | GO:1903284 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.5 | 2.1 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.4 | 1.6 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.3 | 1.9 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.2 | 2.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 1.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.2 | 0.8 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.2 | 0.5 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.1 | 0.9 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.1 | 0.6 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.7 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.1 | 2.7 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.5 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.1 | 0.7 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 0.6 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 0.9 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 1.5 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 1.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.3 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.3 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 1.7 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.9 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 1.1 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.7 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0018963 | phthalate metabolic process(GO:0018963) |
| 0.0 | 0.4 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.6 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.1 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.0 | 2.1 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.7 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.5 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.3 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.3 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 1.2 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.4 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.2 | 1.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.4 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 5.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 1.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.5 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.1 | 0.6 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 5.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.7 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.7 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 3.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 2.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.4 | 5.8 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 1.2 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.2 | 0.8 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.2 | 0.7 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.2 | 3.2 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.2 | 2.7 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 1.6 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.6 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 2.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 2.1 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 1.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 1.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.5 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.5 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 2.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 5.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 2.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 3.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 5.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 3.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 1.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |