Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
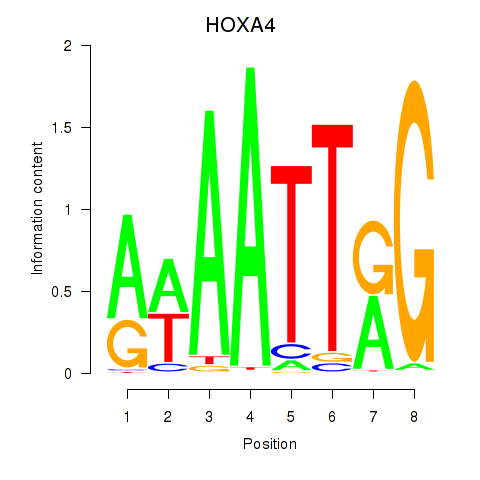
Results for HOXA4
Z-value: 0.43
Transcription factors associated with HOXA4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA4
|
ENSG00000197576.9 | homeobox A4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA4 | hg19_v2_chr7_-_27170352_27170418 | 0.16 | 5.0e-01 | Click! |
Activity profile of HOXA4 motif
Sorted Z-values of HOXA4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_66918558 | 0.50 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr5_-_56778635 | 0.34 |
ENST00000423391.1
|
ACTBL2
|
actin, beta-like 2 |
| chr1_-_100643765 | 0.30 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr9_-_3469181 | 0.29 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr11_+_128563652 | 0.27 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_-_26235206 | 0.25 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr7_+_95115210 | 0.20 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr10_-_105845674 | 0.20 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chrM_+_5824 | 0.19 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr14_-_36988882 | 0.19 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr10_-_105845536 | 0.18 |
ENST00000393211.3
|
COL17A1
|
collagen, type XVII, alpha 1 |
| chr3_+_122103014 | 0.17 |
ENST00000232125.5
ENST00000477892.1 ENST00000469967.1 |
FAM162A
|
family with sequence similarity 162, member A |
| chr6_-_116833500 | 0.17 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr2_-_190927447 | 0.16 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr10_-_75415825 | 0.16 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr6_-_31514333 | 0.16 |
ENST00000376151.4
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr12_+_72058130 | 0.16 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr2_+_171571827 | 0.15 |
ENST00000375281.3
|
SP5
|
Sp5 transcription factor |
| chr3_+_69134124 | 0.15 |
ENST00000478935.1
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr2_-_43453734 | 0.15 |
ENST00000282388.3
|
ZFP36L2
|
ZFP36 ring finger protein-like 2 |
| chr18_-_56985873 | 0.15 |
ENST00000299721.3
|
CPLX4
|
complexin 4 |
| chr1_+_28586006 | 0.14 |
ENST00000253063.3
|
SESN2
|
sestrin 2 |
| chr5_+_49962772 | 0.14 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr19_+_12949251 | 0.14 |
ENST00000251472.4
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr4_-_41884620 | 0.14 |
ENST00000504870.1
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr17_-_38928414 | 0.14 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr3_-_178984759 | 0.14 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr14_-_54423529 | 0.13 |
ENST00000245451.4
ENST00000559087.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr1_+_81771806 | 0.13 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr17_-_67138015 | 0.12 |
ENST00000284425.2
ENST00000590645.1 |
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr11_-_104480019 | 0.12 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr18_+_3247779 | 0.12 |
ENST00000578611.1
ENST00000583449.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr7_-_130598059 | 0.12 |
ENST00000432045.2
|
MIR29B1
|
microRNA 29a |
| chr21_+_17909594 | 0.12 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr14_+_38065052 | 0.12 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr16_+_57728701 | 0.11 |
ENST00000569375.1
ENST00000360716.3 ENST00000569167.1 ENST00000394337.4 ENST00000563126.1 ENST00000336825.8 |
CCDC135
|
coiled-coil domain containing 135 |
| chr18_-_56985776 | 0.11 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr14_-_36989336 | 0.11 |
ENST00000522719.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr7_+_1272522 | 0.11 |
ENST00000316333.8
|
UNCX
|
UNC homeobox |
| chr2_-_163099885 | 0.11 |
ENST00000443424.1
|
FAP
|
fibroblast activation protein, alpha |
| chr2_-_163100045 | 0.11 |
ENST00000188790.4
|
FAP
|
fibroblast activation protein, alpha |
| chr5_+_66300446 | 0.11 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr11_+_128563948 | 0.10 |
ENST00000534087.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr1_+_155658849 | 0.10 |
ENST00000368336.5
ENST00000343043.3 ENST00000421487.2 ENST00000535183.1 ENST00000465375.1 ENST00000470830.1 |
DAP3
|
death associated protein 3 |
| chr11_+_112046190 | 0.10 |
ENST00000357685.5
ENST00000393032.2 ENST00000361053.4 |
BCO2
|
beta-carotene oxygenase 2 |
| chr5_+_66254698 | 0.10 |
ENST00000405643.1
ENST00000407621.1 ENST00000432426.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr20_+_53092123 | 0.10 |
ENST00000262593.5
|
DOK5
|
docking protein 5 |
| chr7_-_27170352 | 0.09 |
ENST00000428284.2
ENST00000360046.5 |
HOXA4
|
homeobox A4 |
| chr14_+_101359265 | 0.09 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr5_-_1882858 | 0.09 |
ENST00000511126.1
ENST00000231357.2 |
IRX4
|
iroquois homeobox 4 |
| chr14_-_38064198 | 0.09 |
ENST00000250448.2
|
FOXA1
|
forkhead box A1 |
| chr14_-_83262540 | 0.09 |
ENST00000554451.1
|
RP11-11K13.1
|
RP11-11K13.1 |
| chr2_-_163099546 | 0.09 |
ENST00000447386.1
|
FAP
|
fibroblast activation protein, alpha |
| chr7_-_143991230 | 0.09 |
ENST00000543357.1
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35 |
| chr10_-_95462265 | 0.09 |
ENST00000536233.1
ENST00000359204.4 ENST00000371430.2 ENST00000394100.2 |
FRA10AC1
|
fragile site, folic acid type, rare, fra(10)(q23.3) or fra(10)(q24.2) candidate 1 |
| chr22_+_26138108 | 0.09 |
ENST00000536101.1
ENST00000335473.7 ENST00000407587.2 |
MYO18B
|
myosin XVIIIB |
| chr19_+_45409011 | 0.09 |
ENST00000252486.4
ENST00000446996.1 ENST00000434152.1 |
APOE
|
apolipoprotein E |
| chr7_-_143892748 | 0.09 |
ENST00000378115.2
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35 |
| chr3_+_52448539 | 0.08 |
ENST00000461861.1
|
PHF7
|
PHD finger protein 7 |
| chr10_-_101825151 | 0.08 |
ENST00000441382.1
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr9_+_44867571 | 0.08 |
ENST00000377548.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr18_+_42260059 | 0.08 |
ENST00000426838.4
|
SETBP1
|
SET binding protein 1 |
| chr7_-_29234802 | 0.08 |
ENST00000449801.1
ENST00000409850.1 |
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr6_-_31514516 | 0.08 |
ENST00000303892.5
ENST00000483251.1 |
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr21_+_17443521 | 0.08 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_+_245318279 | 0.08 |
ENST00000407071.2
|
KIF26B
|
kinesin family member 26B |
| chr20_+_53092232 | 0.08 |
ENST00000395939.1
|
DOK5
|
docking protein 5 |
| chr12_-_16762971 | 0.08 |
ENST00000540590.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr12_+_54447637 | 0.07 |
ENST00000609810.1
ENST00000430889.2 |
HOXC4
HOXC4
|
homeobox C4 Homeobox protein Hox-C4 |
| chr19_-_10047219 | 0.07 |
ENST00000264833.4
|
OLFM2
|
olfactomedin 2 |
| chr2_-_171571077 | 0.07 |
ENST00000409786.1
|
AC007405.2
|
long intergenic non-protein coding RNA 1124 |
| chr4_-_41884582 | 0.07 |
ENST00000499082.2
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr2_-_80531399 | 0.07 |
ENST00000409148.1
ENST00000415098.1 ENST00000452811.1 |
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr7_-_14028488 | 0.07 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
| chr10_+_77056134 | 0.07 |
ENST00000528121.1
ENST00000416398.1 |
ZNF503-AS1
|
ZNF503 antisense RNA 1 |
| chr11_+_22688150 | 0.06 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr6_-_117150198 | 0.06 |
ENST00000310357.3
ENST00000368549.3 ENST00000530250.1 |
GPRC6A
|
G protein-coupled receptor, family C, group 6, member A |
| chr14_+_37131058 | 0.06 |
ENST00000361487.6
|
PAX9
|
paired box 9 |
| chr8_-_139926236 | 0.06 |
ENST00000303045.6
ENST00000435777.1 |
COL22A1
|
collagen, type XXII, alpha 1 |
| chr6_+_31514622 | 0.06 |
ENST00000376146.4
|
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr19_-_1021113 | 0.06 |
ENST00000333175.5
ENST00000356663.3 |
TMEM259
|
transmembrane protein 259 |
| chr21_+_17443434 | 0.06 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr17_-_56082455 | 0.05 |
ENST00000578794.1
|
RP11-159D12.5
|
Uncharacterized protein |
| chr10_+_11047259 | 0.05 |
ENST00000379261.4
ENST00000416382.2 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr19_-_893200 | 0.05 |
ENST00000269814.4
ENST00000395808.3 ENST00000312090.6 ENST00000325464.1 |
MED16
|
mediator complex subunit 16 |
| chr15_+_90744533 | 0.05 |
ENST00000411539.2
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr21_+_22370717 | 0.05 |
ENST00000284894.7
|
NCAM2
|
neural cell adhesion molecule 2 |
| chr20_-_45976816 | 0.05 |
ENST00000441977.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr7_+_141418962 | 0.05 |
ENST00000493845.1
|
WEE2
|
WEE1 homolog 2 (S. pombe) |
| chr4_+_66536248 | 0.05 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr7_-_140714430 | 0.05 |
ENST00000393008.3
|
MRPS33
|
mitochondrial ribosomal protein S33 |
| chr20_+_57464200 | 0.05 |
ENST00000604005.1
|
GNAS
|
GNAS complex locus |
| chr5_+_140810132 | 0.05 |
ENST00000252085.3
|
PCDHGA12
|
protocadherin gamma subfamily A, 12 |
| chr10_+_77056181 | 0.05 |
ENST00000526759.1
ENST00000533822.1 |
ZNF503-AS1
|
ZNF503 antisense RNA 1 |
| chr16_-_85617170 | 0.05 |
ENST00000602862.1
|
RP11-118F19.1
|
RP11-118F19.1 |
| chr5_-_88179302 | 0.05 |
ENST00000504921.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr12_+_12938541 | 0.05 |
ENST00000356591.4
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr2_-_80531824 | 0.05 |
ENST00000295057.3
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr7_-_29235063 | 0.05 |
ENST00000437527.1
ENST00000455544.1 |
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr5_+_49962495 | 0.05 |
ENST00000515175.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr15_+_51669444 | 0.05 |
ENST00000396399.2
|
GLDN
|
gliomedin |
| chr3_+_88188254 | 0.05 |
ENST00000309495.5
|
ZNF654
|
zinc finger protein 654 |
| chr11_+_112832133 | 0.05 |
ENST00000524665.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr3_-_64253655 | 0.05 |
ENST00000498162.1
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr1_+_151739131 | 0.05 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chrX_-_55024967 | 0.04 |
ENST00000545676.1
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chrX_+_134654540 | 0.04 |
ENST00000370752.4
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr5_-_13944652 | 0.04 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr17_-_10452929 | 0.04 |
ENST00000532183.2
ENST00000397183.2 ENST00000420805.1 |
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult |
| chr14_+_39734482 | 0.04 |
ENST00000554392.1
ENST00000555716.1 ENST00000341749.3 ENST00000557038.1 |
CTAGE5
|
CTAGE family, member 5 |
| chr5_+_69321074 | 0.04 |
ENST00000380751.5
ENST00000380750.3 ENST00000503931.1 ENST00000506542.1 |
SERF1B
|
small EDRK-rich factor 1B (centromeric) |
| chr5_+_133562095 | 0.04 |
ENST00000602919.1
|
CTD-2410N18.3
|
CTD-2410N18.3 |
| chr1_-_12677714 | 0.04 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr21_+_17442799 | 0.04 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr9_-_16253112 | 0.04 |
ENST00000380683.1
|
C9orf92
|
chromosome 9 open reading frame 92 |
| chr12_-_68845165 | 0.04 |
ENST00000360485.3
ENST00000441255.2 |
RP11-81H14.2
|
RP11-81H14.2 |
| chr2_+_113825547 | 0.04 |
ENST00000341010.2
ENST00000337569.3 |
IL1F10
|
interleukin 1 family, member 10 (theta) |
| chr8_-_99954788 | 0.04 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr6_-_55740352 | 0.04 |
ENST00000370830.3
|
BMP5
|
bone morphogenetic protein 5 |
| chr7_+_18330035 | 0.04 |
ENST00000413509.2
|
HDAC9
|
histone deacetylase 9 |
| chr12_-_24103954 | 0.04 |
ENST00000441133.2
ENST00000545921.1 |
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr12_-_11091862 | 0.03 |
ENST00000537503.1
|
TAS2R14
|
taste receptor, type 2, member 14 |
| chr1_+_214161272 | 0.03 |
ENST00000498508.2
ENST00000366958.4 |
PROX1
|
prospero homeobox 1 |
| chr2_+_39117010 | 0.03 |
ENST00000409978.1
|
ARHGEF33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr4_-_66536057 | 0.03 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr18_-_53019208 | 0.03 |
ENST00000562607.1
|
TCF4
|
transcription factor 4 |
| chr9_+_87284622 | 0.03 |
ENST00000395882.1
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chrX_-_119445263 | 0.03 |
ENST00000309720.5
|
TMEM255A
|
transmembrane protein 255A |
| chr6_+_50786414 | 0.03 |
ENST00000344788.3
ENST00000393655.3 ENST00000263046.4 |
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta) |
| chr12_-_57444957 | 0.03 |
ENST00000433964.1
|
MYO1A
|
myosin IA |
| chr8_+_39792474 | 0.03 |
ENST00000502986.2
|
IDO2
|
indoleamine 2,3-dioxygenase 2 |
| chr4_-_109541610 | 0.03 |
ENST00000510212.1
|
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr4_-_109541539 | 0.03 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chrX_+_37639302 | 0.03 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr9_-_113341985 | 0.03 |
ENST00000374469.1
|
SVEP1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr21_+_17961006 | 0.03 |
ENST00000602323.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr17_-_10421853 | 0.03 |
ENST00000226207.5
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult |
| chr4_+_189060573 | 0.03 |
ENST00000332517.3
|
TRIML1
|
tripartite motif family-like 1 |
| chr2_-_180427304 | 0.03 |
ENST00000336917.5
|
ZNF385B
|
zinc finger protein 385B |
| chr1_+_160160283 | 0.03 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr15_+_80351977 | 0.03 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr11_+_120971882 | 0.03 |
ENST00000392793.1
|
TECTA
|
tectorin alpha |
| chr11_+_4470525 | 0.03 |
ENST00000325719.4
|
OR52K2
|
olfactory receptor, family 52, subfamily K, member 2 |
| chr15_+_51669513 | 0.03 |
ENST00000558426.1
|
GLDN
|
gliomedin |
| chr9_+_87284675 | 0.02 |
ENST00000376208.1
ENST00000304053.6 ENST00000277120.3 |
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr15_-_33447055 | 0.02 |
ENST00000559047.1
ENST00000561249.1 |
FMN1
|
formin 1 |
| chr4_-_185139062 | 0.02 |
ENST00000296741.2
|
ENPP6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr19_-_51220176 | 0.02 |
ENST00000359082.3
ENST00000293441.1 |
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr5_+_147582348 | 0.02 |
ENST00000514389.1
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr6_+_152011628 | 0.02 |
ENST00000404742.1
ENST00000440973.1 |
ESR1
|
estrogen receptor 1 |
| chr5_+_152870734 | 0.02 |
ENST00000521843.2
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1 |
| chr13_-_46716969 | 0.02 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr8_+_71383312 | 0.02 |
ENST00000520259.1
|
RP11-333A23.4
|
RP11-333A23.4 |
| chr12_-_86650045 | 0.02 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr1_+_167298281 | 0.02 |
ENST00000367862.5
|
POU2F1
|
POU class 2 homeobox 1 |
| chr7_-_14026123 | 0.02 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr9_+_75263565 | 0.02 |
ENST00000396237.3
|
TMC1
|
transmembrane channel-like 1 |
| chr18_-_70532906 | 0.02 |
ENST00000299430.2
ENST00000397929.1 |
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr3_+_159557637 | 0.02 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr12_+_111537227 | 0.02 |
ENST00000397643.3
|
CUX2
|
cut-like homeobox 2 |
| chr3_-_57233966 | 0.02 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr8_+_77593448 | 0.02 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr3_+_121311966 | 0.02 |
ENST00000338040.4
|
FBXO40
|
F-box protein 40 |
| chrX_+_37639264 | 0.02 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr6_+_63921351 | 0.02 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr3_-_42744270 | 0.02 |
ENST00000457462.1
|
HHATL
|
hedgehog acyltransferase-like |
| chr1_+_160160346 | 0.02 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr5_+_152870287 | 0.02 |
ENST00000340592.5
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1 |
| chr5_+_152870215 | 0.01 |
ENST00000518142.1
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1 |
| chr9_+_26746951 | 0.01 |
ENST00000523363.1
|
RP11-18A15.1
|
RP11-18A15.1 |
| chr12_+_29376592 | 0.01 |
ENST00000182377.4
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr7_+_117251671 | 0.01 |
ENST00000468795.1
|
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chrX_+_41548220 | 0.01 |
ENST00000378142.4
|
GPR34
|
G protein-coupled receptor 34 |
| chr5_+_60933634 | 0.01 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr4_-_66536196 | 0.01 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chrX_+_41548259 | 0.01 |
ENST00000378138.5
|
GPR34
|
G protein-coupled receptor 34 |
| chr5_+_115387163 | 0.01 |
ENST00000601302.2
|
ARL14EPL
|
ADP-ribosylation factor-like 14 effector protein-like |
| chr18_+_28956740 | 0.01 |
ENST00000308128.4
ENST00000359747.4 |
DSG4
|
desmoglein 4 |
| chrX_-_119445306 | 0.01 |
ENST00000371369.4
ENST00000440464.1 ENST00000519908.1 |
TMEM255A
|
transmembrane protein 255A |
| chr5_+_147582387 | 0.01 |
ENST00000325630.2
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr3_+_69134080 | 0.01 |
ENST00000273258.3
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr12_+_29376673 | 0.01 |
ENST00000547116.1
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr15_+_81299370 | 0.01 |
ENST00000560091.1
|
C15orf26
|
chromosome 15 open reading frame 26 |
| chr17_+_37894570 | 0.01 |
ENST00000394211.3
|
GRB7
|
growth factor receptor-bound protein 7 |
| chr20_-_45035223 | 0.01 |
ENST00000450812.1
ENST00000290246.6 ENST00000439931.2 ENST00000396391.1 |
ELMO2
|
engulfment and cell motility 2 |
| chr10_-_106240032 | 0.01 |
ENST00000447860.1
|
RP11-127O4.3
|
RP11-127O4.3 |
| chr5_+_66300464 | 0.01 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr12_+_106976678 | 0.01 |
ENST00000392842.1
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr1_-_108231101 | 0.01 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr11_+_124481361 | 0.01 |
ENST00000284288.2
|
PANX3
|
pannexin 3 |
| chrX_+_129473916 | 0.01 |
ENST00000545805.1
ENST00000543953.1 ENST00000218197.5 |
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr17_-_39211463 | 0.01 |
ENST00000542910.1
ENST00000398477.1 |
KRTAP2-2
|
keratin associated protein 2-2 |
| chr8_+_77593474 | 0.01 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr8_-_10512569 | 0.00 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr2_+_17997763 | 0.00 |
ENST00000281047.3
|
MSGN1
|
mesogenin 1 |
| chr9_-_16276311 | 0.00 |
ENST00000380685.1
|
C9orf92
|
chromosome 9 open reading frame 92 |
| chr18_-_28742813 | 0.00 |
ENST00000257197.3
ENST00000257198.5 |
DSC1
|
desmocollin 1 |
| chr10_-_62332357 | 0.00 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr9_-_113342160 | 0.00 |
ENST00000401783.2
ENST00000374461.1 |
SVEP1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr1_-_216978709 | 0.00 |
ENST00000360012.3
|
ESRRG
|
estrogen-related receptor gamma |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1902362 | melanocyte apoptotic process(GO:1902362) |
| 0.0 | 0.2 | GO:1902725 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.0 | 0.1 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.0 | 0.1 | GO:2000646 | lipid transport involved in lipid storage(GO:0010877) positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0071893 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.2 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.1 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.0 | GO:1905069 | allantois development(GO:1905069) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.2 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.0 | GO:2000979 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 | 0.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.3 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |