Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
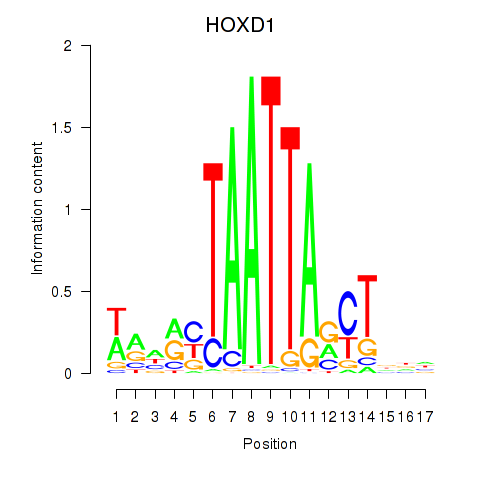
Results for HOXD1
Z-value: 0.59
Transcription factors associated with HOXD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD1
|
ENSG00000128645.11 | homeobox D1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD1 | hg19_v2_chr2_+_177053307_177053402 | -0.58 | 6.7e-03 | Click! |
Activity profile of HOXD1 motif
Sorted Z-values of HOXD1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_151034734 | 2.52 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chrX_+_43515467 | 2.34 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr8_-_49834299 | 2.32 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr8_-_49833978 | 2.14 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr19_+_11071546 | 1.99 |
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr11_-_102651343 | 1.82 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr11_+_65554493 | 1.67 |
ENST00000335987.3
|
OVOL1
|
ovo-like zinc finger 1 |
| chr3_+_111717511 | 1.41 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr5_+_66300464 | 1.38 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr11_+_71900703 | 1.37 |
ENST00000393681.2
|
FOLR1
|
folate receptor 1 (adult) |
| chr11_+_71900572 | 1.29 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
| chr4_-_74486217 | 1.24 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr5_+_66300446 | 1.17 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr3_+_111718036 | 1.16 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr4_+_144354644 | 1.08 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr3_+_111717600 | 1.05 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr4_-_74486347 | 1.03 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr4_-_74486109 | 1.01 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr19_-_36001113 | 0.94 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr6_+_130339710 | 0.88 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr4_+_144312659 | 0.81 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr9_+_124329336 | 0.81 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr3_-_99569821 | 0.78 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr13_-_36050819 | 0.68 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr18_-_33709268 | 0.65 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr3_+_111718173 | 0.63 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr11_-_26593677 | 0.61 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr4_-_119759795 | 0.59 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr1_+_117544366 | 0.58 |
ENST00000256652.4
ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr11_-_26593779 | 0.57 |
ENST00000529533.1
|
MUC15
|
mucin 15, cell surface associated |
| chr12_-_22063787 | 0.55 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr12_-_10022735 | 0.53 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr9_-_5339873 | 0.51 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr10_+_99205894 | 0.47 |
ENST00000370854.3
ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr4_-_25865159 | 0.44 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr11_-_26593649 | 0.40 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr5_+_67535647 | 0.38 |
ENST00000520675.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr14_+_32798462 | 0.38 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr2_+_191221240 | 0.37 |
ENST00000409027.1
ENST00000458193.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr10_+_99205959 | 0.36 |
ENST00000352634.4
ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr14_+_57671888 | 0.33 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chrX_+_107288280 | 0.33 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr5_+_148737562 | 0.32 |
ENST00000274569.4
|
PCYOX1L
|
prenylcysteine oxidase 1 like |
| chr16_+_53133070 | 0.31 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr15_-_55562451 | 0.30 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr17_+_18086392 | 0.29 |
ENST00000541285.1
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli) |
| chr7_+_37723450 | 0.27 |
ENST00000447769.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr7_+_37723336 | 0.27 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr7_+_133812052 | 0.26 |
ENST00000285928.2
|
LRGUK
|
leucine-rich repeats and guanylate kinase domain containing |
| chr4_+_86749045 | 0.18 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr3_-_126327398 | 0.18 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr12_-_118797475 | 0.17 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr4_-_69111401 | 0.15 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr1_-_151431674 | 0.14 |
ENST00000531094.1
|
POGZ
|
pogo transposable element with ZNF domain |
| chrX_+_107288239 | 0.14 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr12_+_29376592 | 0.13 |
ENST00000182377.4
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr11_-_71823715 | 0.13 |
ENST00000545944.1
ENST00000502597.2 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr12_+_64798826 | 0.13 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr5_+_63802109 | 0.13 |
ENST00000334025.2
|
RGS7BP
|
regulator of G-protein signaling 7 binding protein |
| chr11_-_71823796 | 0.12 |
ENST00000545680.1
ENST00000543587.1 ENST00000538393.1 ENST00000535234.1 ENST00000227618.4 ENST00000535503.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr12_+_29376673 | 0.11 |
ENST00000547116.1
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr11_+_4615269 | 0.11 |
ENST00000530443.2
|
OR52I1
|
olfactory receptor, family 52, subfamily I, member 1 |
| chr5_+_36606700 | 0.10 |
ENST00000416645.2
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr4_+_86525299 | 0.10 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr11_-_62521614 | 0.09 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr2_+_228736335 | 0.09 |
ENST00000440997.1
ENST00000545118.1 |
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr22_-_24951888 | 0.09 |
ENST00000404664.3
|
GUCD1
|
guanylyl cyclase domain containing 1 |
| chr11_+_4615037 | 0.08 |
ENST00000450052.2
|
OR52I1
|
olfactory receptor, family 52, subfamily I, member 1 |
| chr14_+_32798547 | 0.07 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr7_-_81399329 | 0.07 |
ENST00000453411.1
ENST00000444829.2 |
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr1_-_151431647 | 0.07 |
ENST00000368863.2
ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ
|
pogo transposable element with ZNF domain |
| chr7_-_81399355 | 0.06 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chrX_+_107288197 | 0.06 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr5_+_176811431 | 0.06 |
ENST00000512593.1
ENST00000324417.5 |
SLC34A1
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 1 |
| chr11_-_18211042 | 0.05 |
ENST00000527671.1
|
RP11-113D6.6
|
Uncharacterized protein |
| chr10_-_4285923 | 0.05 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr1_-_92952433 | 0.04 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr4_-_139051839 | 0.04 |
ENST00000514600.1
ENST00000513895.1 ENST00000512536.1 |
LINC00616
|
long intergenic non-protein coding RNA 616 |
| chr11_-_71823266 | 0.03 |
ENST00000538919.1
ENST00000539395.1 ENST00000542531.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr12_+_10163231 | 0.02 |
ENST00000396502.1
ENST00000338896.5 |
CLEC12B
|
C-type lectin domain family 12, member B |
| chr3_-_101039402 | 0.02 |
ENST00000193391.7
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr7_-_81399287 | 0.02 |
ENST00000354224.6
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr7_-_81399411 | 0.02 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr2_+_54350316 | 0.01 |
ENST00000606865.1
|
ACYP2
|
acylphosphatase 2, muscle type |
| chr12_+_15699286 | 0.01 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr10_-_4285835 | 0.01 |
ENST00000454470.1
|
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr4_+_86748898 | 0.01 |
ENST00000509300.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr2_+_135596106 | 0.00 |
ENST00000356140.5
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr4_-_150736962 | 0.00 |
ENST00000502345.1
ENST00000510975.1 ENST00000511993.1 |
RP11-526A4.1
|
RP11-526A4.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.9 | 2.7 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.4 | 1.7 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.4 | 2.0 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.2 | 0.8 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 2.3 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.9 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.3 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 2.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.3 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 2.0 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.2 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 1.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 1.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.4 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.0 | GO:0070105 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.3 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.1 | 2.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 2.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.6 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.4 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.3 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 4.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.6 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.3 | 2.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.6 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.8 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 2.0 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 2.5 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.4 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.1 | 0.6 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 0.3 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.3 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 2.3 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 6.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 6.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 2.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |