Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
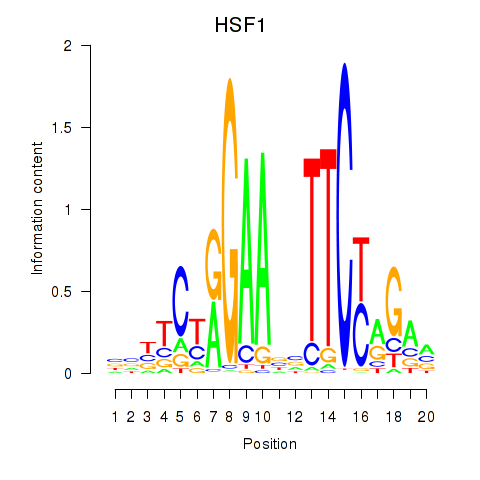
Results for HSF1
Z-value: 0.50
Transcription factors associated with HSF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF1
|
ENSG00000185122.6 | heat shock transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSF1 | hg19_v2_chr8_+_145515263_145515299 | -0.62 | 3.6e-03 | Click! |
Activity profile of HSF1 motif
Sorted Z-values of HSF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_52911718 | 2.24 |
ENST00000548409.1
|
KRT5
|
keratin 5 |
| chr17_-_39661849 | 1.75 |
ENST00000246635.3
ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13
|
keratin 13 |
| chr16_+_56659687 | 1.30 |
ENST00000568293.1
ENST00000330439.6 |
MT1E
|
metallothionein 1E |
| chr1_-_153363452 | 1.22 |
ENST00000368732.1
ENST00000368733.3 |
S100A8
|
S100 calcium binding protein A8 |
| chr17_-_39928106 | 1.22 |
ENST00000540235.1
|
JUP
|
junction plakoglobin |
| chr11_-_104840093 | 1.15 |
ENST00000417440.2
ENST00000444739.2 |
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chrX_-_73072534 | 1.14 |
ENST00000429829.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr17_-_39661947 | 1.14 |
ENST00000590425.1
|
KRT13
|
keratin 13 |
| chr8_+_124194752 | 1.13 |
ENST00000318462.6
|
FAM83A
|
family with sequence similarity 83, member A |
| chr1_+_35247859 | 1.13 |
ENST00000373362.3
|
GJB3
|
gap junction protein, beta 3, 31kDa |
| chr2_+_237476419 | 0.99 |
ENST00000447924.1
|
ACKR3
|
atypical chemokine receptor 3 |
| chr7_+_157129738 | 0.94 |
ENST00000437030.1
|
DNAJB6
|
DnaJ (Hsp40) homolog, subfamily B, member 6 |
| chr17_-_39677971 | 0.91 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr6_+_35265586 | 0.85 |
ENST00000542066.1
ENST00000316637.5 |
DEF6
|
differentially expressed in FDCP 6 homolog (mouse) |
| chr11_-_104905840 | 0.81 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr6_+_47666275 | 0.79 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr19_-_36004543 | 0.75 |
ENST00000339686.3
ENST00000447113.2 ENST00000440396.1 |
DMKN
|
dermokine |
| chr2_+_113735575 | 0.74 |
ENST00000376489.2
ENST00000259205.4 |
IL36G
|
interleukin 36, gamma |
| chr19_-_51522955 | 0.71 |
ENST00000358789.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr7_-_3083573 | 0.70 |
ENST00000396946.4
|
CARD11
|
caspase recruitment domain family, member 11 |
| chr7_+_150020363 | 0.65 |
ENST00000359623.4
ENST00000493307.1 |
LRRC61
|
leucine rich repeat containing 61 |
| chr2_-_56150910 | 0.64 |
ENST00000424836.2
ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr11_+_123986069 | 0.64 |
ENST00000456829.2
ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A
|
von Willebrand factor A domain containing 5A |
| chr11_+_34645791 | 0.62 |
ENST00000529527.1
ENST00000531728.1 ENST00000525253.1 |
EHF
|
ets homologous factor |
| chr19_-_51456344 | 0.62 |
ENST00000336334.3
ENST00000593428.1 |
KLK5
|
kallikrein-related peptidase 5 |
| chr18_+_21452804 | 0.60 |
ENST00000269217.6
|
LAMA3
|
laminin, alpha 3 |
| chr19_-_53193731 | 0.59 |
ENST00000598536.1
ENST00000594682.2 ENST00000601257.1 |
ZNF83
|
zinc finger protein 83 |
| chr19_-_12444306 | 0.59 |
ENST00000594577.1
ENST00000601858.1 |
ZNF563
|
zinc finger protein 563 |
| chr14_+_24837226 | 0.58 |
ENST00000554050.1
ENST00000554903.1 ENST00000554779.1 ENST00000250373.4 ENST00000553708.1 |
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
| chr19_-_51523412 | 0.57 |
ENST00000391805.1
ENST00000599077.1 |
KLK10
|
kallikrein-related peptidase 10 |
| chr1_+_19970202 | 0.57 |
ENST00000439664.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr14_-_24809154 | 0.57 |
ENST00000216274.5
|
RIPK3
|
receptor-interacting serine-threonine kinase 3 |
| chr18_+_21452964 | 0.57 |
ENST00000587184.1
|
LAMA3
|
laminin, alpha 3 |
| chr6_-_32784687 | 0.56 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr19_-_6720686 | 0.54 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr6_-_170599561 | 0.54 |
ENST00000366756.3
|
DLL1
|
delta-like 1 (Drosophila) |
| chr4_-_77819002 | 0.53 |
ENST00000334306.2
|
SOWAHB
|
sosondowah ankyrin repeat domain family member B |
| chr16_-_79634595 | 0.53 |
ENST00000326043.4
ENST00000393350.1 |
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr6_+_46620676 | 0.52 |
ENST00000371347.5
ENST00000411689.2 |
SLC25A27
|
solute carrier family 25, member 27 |
| chr7_+_150020329 | 0.51 |
ENST00000323078.7
|
LRRC61
|
leucine rich repeat containing 61 |
| chr19_-_2051223 | 0.50 |
ENST00000309340.7
ENST00000589534.1 ENST00000250896.3 ENST00000589509.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr19_-_51456198 | 0.50 |
ENST00000594846.1
|
KLK5
|
kallikrein-related peptidase 5 |
| chr19_-_51523275 | 0.50 |
ENST00000309958.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr4_+_106816644 | 0.49 |
ENST00000506666.1
ENST00000503451.1 |
NPNT
|
nephronectin |
| chr1_-_27682962 | 0.49 |
ENST00000486046.1
|
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr12_-_123011476 | 0.49 |
ENST00000528279.1
ENST00000344591.4 ENST00000526560.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chrX_+_147062844 | 0.48 |
ENST00000370467.3
|
FMR1NB
|
fragile X mental retardation 1 neighbor |
| chr19_-_2085323 | 0.48 |
ENST00000591638.1
|
MOB3A
|
MOB kinase activator 3A |
| chr15_-_72490114 | 0.47 |
ENST00000309731.7
|
GRAMD2
|
GRAM domain containing 2 |
| chr1_-_183538319 | 0.47 |
ENST00000420553.1
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr1_+_10003486 | 0.46 |
ENST00000403197.1
ENST00000377205.1 |
NMNAT1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr1_+_161494036 | 0.45 |
ENST00000309758.4
|
HSPA6
|
heat shock 70kDa protein 6 (HSP70B') |
| chr11_-_111782696 | 0.44 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr17_+_8339340 | 0.44 |
ENST00000580012.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr11_-_125773085 | 0.44 |
ENST00000227474.3
ENST00000534158.1 ENST00000529801.1 |
PUS3
|
pseudouridylate synthase 3 |
| chr2_-_220142892 | 0.44 |
ENST00000427737.1
|
TUBA4A
|
tubulin, alpha 4a |
| chr2_-_241831424 | 0.44 |
ENST00000402775.2
ENST00000307486.8 |
C2orf54
|
chromosome 2 open reading frame 54 |
| chr21_+_35445827 | 0.43 |
ENST00000608209.1
ENST00000381151.3 |
SLC5A3
SLC5A3
|
sodium/myo-inositol cotransporter solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
| chr2_+_47596287 | 0.41 |
ENST00000263735.4
|
EPCAM
|
epithelial cell adhesion molecule |
| chr11_+_10471836 | 0.41 |
ENST00000444303.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr20_-_62168714 | 0.40 |
ENST00000542869.1
|
PTK6
|
protein tyrosine kinase 6 |
| chr6_-_38607628 | 0.40 |
ENST00000498633.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr2_-_151395525 | 0.40 |
ENST00000439275.1
|
RND3
|
Rho family GTPase 3 |
| chr19_+_8429031 | 0.40 |
ENST00000301455.2
ENST00000541807.1 ENST00000393962.2 |
ANGPTL4
|
angiopoietin-like 4 |
| chr12_+_119772502 | 0.39 |
ENST00000536742.1
ENST00000327554.2 |
CCDC60
|
coiled-coil domain containing 60 |
| chr3_+_44916098 | 0.39 |
ENST00000296125.4
|
TGM4
|
transglutaminase 4 |
| chr21_+_39644172 | 0.39 |
ENST00000398932.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr6_+_46620705 | 0.39 |
ENST00000452689.2
|
SLC25A27
|
solute carrier family 25, member 27 |
| chr22_-_36556821 | 0.38 |
ENST00000531095.1
ENST00000397293.2 ENST00000349314.2 |
APOL3
|
apolipoprotein L, 3 |
| chr6_+_32937083 | 0.38 |
ENST00000456339.1
|
BRD2
|
bromodomain containing 2 |
| chr19_-_10628117 | 0.38 |
ENST00000333430.4
|
S1PR5
|
sphingosine-1-phosphate receptor 5 |
| chr9_+_71736177 | 0.38 |
ENST00000606364.1
ENST00000453658.2 |
TJP2
|
tight junction protein 2 |
| chr14_+_101293687 | 0.37 |
ENST00000455286.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr14_+_75988768 | 0.36 |
ENST00000286639.6
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr20_-_62168672 | 0.36 |
ENST00000217185.2
|
PTK6
|
protein tyrosine kinase 6 |
| chr21_+_39644214 | 0.36 |
ENST00000438657.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr1_-_109584716 | 0.36 |
ENST00000531337.1
ENST00000529074.1 ENST00000369965.4 |
WDR47
|
WD repeat domain 47 |
| chr17_+_4901199 | 0.36 |
ENST00000320785.5
ENST00000574165.1 |
KIF1C
|
kinesin family member 1C |
| chrX_+_37208540 | 0.35 |
ENST00000466533.1
ENST00000542554.1 ENST00000543642.1 ENST00000484460.1 ENST00000449135.2 ENST00000463135.1 ENST00000465127.1 |
PRRG1
TM4SF2
|
proline rich Gla (G-carboxyglutamic acid) 1 Uncharacterized protein; cDNA FLJ59144, highly similar to Tetraspanin-7 |
| chr20_+_43803517 | 0.35 |
ENST00000243924.3
|
PI3
|
peptidase inhibitor 3, skin-derived |
| chr6_-_10419871 | 0.34 |
ENST00000319516.4
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr3_+_156393349 | 0.34 |
ENST00000473702.1
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr3_-_122233723 | 0.34 |
ENST00000493510.1
ENST00000344337.6 ENST00000476916.1 ENST00000465882.1 |
KPNA1
|
karyopherin alpha 1 (importin alpha 5) |
| chr19_-_14629224 | 0.34 |
ENST00000254322.2
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr2_-_238499131 | 0.33 |
ENST00000538644.1
|
RAB17
|
RAB17, member RAS oncogene family |
| chr3_-_183735731 | 0.33 |
ENST00000334444.6
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr3_+_38032216 | 0.33 |
ENST00000416303.1
|
VILL
|
villin-like |
| chr20_+_37230511 | 0.33 |
ENST00000373345.4
|
ARHGAP40
|
Rho GTPase activating protein 40 |
| chr6_-_29648887 | 0.32 |
ENST00000376883.1
|
ZFP57
|
ZFP57 zinc finger protein |
| chr19_+_56905024 | 0.32 |
ENST00000591172.1
ENST00000589888.1 ENST00000587979.1 ENST00000585659.1 ENST00000593109.1 |
ZNF582-AS1
|
ZNF582 antisense RNA 1 (head to head) |
| chr12_-_10826612 | 0.32 |
ENST00000535345.1
ENST00000542562.1 |
STYK1
|
serine/threonine/tyrosine kinase 1 |
| chr3_+_42977846 | 0.32 |
ENST00000383748.4
|
KRBOX1
|
KRAB box domain containing 1 |
| chr22_-_38699003 | 0.32 |
ENST00000451964.1
|
CSNK1E
|
casein kinase 1, epsilon |
| chrX_-_7895755 | 0.31 |
ENST00000444736.1
ENST00000537427.1 ENST00000442940.1 |
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr1_-_109584768 | 0.31 |
ENST00000357672.3
|
WDR47
|
WD repeat domain 47 |
| chr19_-_49567124 | 0.31 |
ENST00000301411.3
|
NTF4
|
neurotrophin 4 |
| chr18_+_14748194 | 0.31 |
ENST00000358984.4
|
ANKRD30B
|
ankyrin repeat domain 30B |
| chr1_-_27709816 | 0.31 |
ENST00000374030.1
|
CD164L2
|
CD164 sialomucin-like 2 |
| chr7_-_150020750 | 0.31 |
ENST00000539352.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr14_+_71165292 | 0.30 |
ENST00000553682.1
|
RP6-65G23.1
|
RP6-65G23.1 |
| chr1_-_116926718 | 0.30 |
ENST00000598661.1
|
AL136376.1
|
Uncharacterized protein |
| chr7_+_157129660 | 0.30 |
ENST00000429029.2
ENST00000262177.4 ENST00000417758.1 ENST00000452797.2 ENST00000443280.1 |
DNAJB6
|
DnaJ (Hsp40) homolog, subfamily B, member 6 |
| chr19_-_2151523 | 0.30 |
ENST00000350812.6
ENST00000355272.6 ENST00000356926.4 ENST00000345016.5 |
AP3D1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr14_-_21493123 | 0.29 |
ENST00000556147.1
ENST00000554489.1 ENST00000555657.1 ENST00000557274.1 ENST00000555158.1 ENST00000554833.1 ENST00000555384.1 ENST00000556420.1 ENST00000554893.1 ENST00000553503.1 ENST00000555733.1 ENST00000553867.1 ENST00000397856.3 ENST00000397855.3 ENST00000556008.1 ENST00000557182.1 ENST00000554483.1 ENST00000556688.1 ENST00000397853.3 ENST00000556329.2 ENST00000554143.1 ENST00000397851.2 ENST00000555142.1 ENST00000557676.1 ENST00000556924.1 |
NDRG2
|
NDRG family member 2 |
| chr11_+_64323098 | 0.29 |
ENST00000301891.4
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr5_+_149887672 | 0.29 |
ENST00000261797.6
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr21_+_42733870 | 0.29 |
ENST00000330714.3
ENST00000436410.1 ENST00000435611.1 |
MX2
|
myxovirus (influenza virus) resistance 2 (mouse) |
| chr9_+_470288 | 0.29 |
ENST00000382303.1
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr12_-_47219733 | 0.29 |
ENST00000547477.1
ENST00000447411.1 ENST00000266579.4 |
SLC38A4
|
solute carrier family 38, member 4 |
| chr11_+_33563821 | 0.28 |
ENST00000321505.4
ENST00000265654.5 ENST00000389726.3 |
KIAA1549L
|
KIAA1549-like |
| chrX_+_150866779 | 0.28 |
ENST00000370353.3
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr1_-_47082495 | 0.28 |
ENST00000545730.1
ENST00000531769.1 ENST00000319928.3 |
MKNK1
MOB3C
|
MAP kinase interacting serine/threonine kinase 1 MOB kinase activator 3C |
| chr8_-_8318847 | 0.28 |
ENST00000521218.1
|
CTA-398F10.2
|
CTA-398F10.2 |
| chr2_-_9143786 | 0.28 |
ENST00000462696.1
ENST00000305997.3 |
MBOAT2
|
membrane bound O-acyltransferase domain containing 2 |
| chr6_-_46620522 | 0.28 |
ENST00000275016.2
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr7_-_150020814 | 0.28 |
ENST00000477871.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr2_+_28718921 | 0.28 |
ENST00000327757.5
ENST00000422425.2 ENST00000404858.1 |
PLB1
|
phospholipase B1 |
| chr20_-_18038521 | 0.27 |
ENST00000278780.6
|
OVOL2
|
ovo-like zinc finger 2 |
| chr9_+_72658490 | 0.27 |
ENST00000377182.4
|
MAMDC2
|
MAM domain containing 2 |
| chr4_+_57396766 | 0.27 |
ENST00000512175.2
|
THEGL
|
theg spermatid protein-like |
| chr11_+_65851443 | 0.27 |
ENST00000533756.1
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr3_-_18487057 | 0.27 |
ENST00000415069.1
|
SATB1
|
SATB homeobox 1 |
| chr4_+_119200215 | 0.26 |
ENST00000602573.1
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr4_+_166300084 | 0.26 |
ENST00000402744.4
|
CPE
|
carboxypeptidase E |
| chr19_-_14628234 | 0.26 |
ENST00000595139.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr6_+_29910301 | 0.26 |
ENST00000376809.5
ENST00000376802.2 |
HLA-A
|
major histocompatibility complex, class I, A |
| chr10_+_24528108 | 0.26 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr12_-_10875831 | 0.26 |
ENST00000279550.7
ENST00000228251.4 |
YBX3
|
Y box binding protein 3 |
| chr11_+_33563618 | 0.26 |
ENST00000526400.1
|
KIAA1549L
|
KIAA1549-like |
| chr10_-_90611566 | 0.26 |
ENST00000371930.4
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr11_-_111782484 | 0.26 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr20_+_58203664 | 0.26 |
ENST00000541461.1
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr3_+_42947600 | 0.26 |
ENST00000328199.6
ENST00000541208.1 |
ZNF662
|
zinc finger protein 662 |
| chr20_+_1875378 | 0.26 |
ENST00000356025.3
|
SIRPA
|
signal-regulatory protein alpha |
| chr7_-_139763521 | 0.25 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr10_+_15001430 | 0.25 |
ENST00000407572.1
|
MEIG1
|
meiosis/spermiogenesis associated 1 |
| chr2_+_220144052 | 0.25 |
ENST00000425450.1
ENST00000392086.4 ENST00000421532.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr17_-_43138357 | 0.25 |
ENST00000342350.5
|
DCAKD
|
dephospho-CoA kinase domain containing |
| chr11_-_914812 | 0.25 |
ENST00000533059.1
|
CHID1
|
chitinase domain containing 1 |
| chr19_-_56904799 | 0.25 |
ENST00000589895.1
ENST00000589143.1 ENST00000301310.4 ENST00000586929.1 |
ZNF582
|
zinc finger protein 582 |
| chr19_-_44952635 | 0.25 |
ENST00000592308.1
ENST00000588931.1 ENST00000291187.4 |
ZNF229
|
zinc finger protein 229 |
| chr11_-_134123142 | 0.25 |
ENST00000392595.2
ENST00000341541.3 ENST00000352327.5 ENST00000392594.3 |
THYN1
|
thymocyte nuclear protein 1 |
| chr18_-_32870148 | 0.25 |
ENST00000589178.1
ENST00000333206.5 ENST00000592278.1 ENST00000592211.1 ENST00000420878.3 ENST00000383091.2 ENST00000586922.2 |
ZSCAN30
RP11-158H5.7
|
zinc finger and SCAN domain containing 30 RP11-158H5.7 |
| chr11_-_33744256 | 0.24 |
ENST00000415002.2
ENST00000437761.2 ENST00000445143.2 |
CD59
|
CD59 molecule, complement regulatory protein |
| chr13_-_79979919 | 0.24 |
ENST00000267229.7
|
RBM26
|
RNA binding motif protein 26 |
| chr13_-_79979952 | 0.24 |
ENST00000438724.1
|
RBM26
|
RNA binding motif protein 26 |
| chr19_-_12444519 | 0.24 |
ENST00000595977.1
|
ZNF563
|
zinc finger protein 563 |
| chr2_+_220143989 | 0.24 |
ENST00000336576.5
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr6_+_10521574 | 0.24 |
ENST00000495262.1
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chrX_+_37208521 | 0.24 |
ENST00000378628.4
|
PRRG1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr3_-_59035673 | 0.24 |
ENST00000491845.1
ENST00000472469.1 ENST00000471288.1 ENST00000295966.7 |
C3orf67
|
chromosome 3 open reading frame 67 |
| chr1_-_236030216 | 0.24 |
ENST00000389794.3
ENST00000389793.2 |
LYST
|
lysosomal trafficking regulator |
| chr11_-_88070896 | 0.24 |
ENST00000529974.1
ENST00000527018.1 |
CTSC
|
cathepsin C |
| chr12_-_123011536 | 0.24 |
ENST00000331738.7
ENST00000354654.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr9_+_109685630 | 0.23 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr1_-_27709793 | 0.23 |
ENST00000374027.3
ENST00000374025.3 |
CD164L2
|
CD164 sialomucin-like 2 |
| chr1_+_152957707 | 0.23 |
ENST00000368762.1
|
SPRR1A
|
small proline-rich protein 1A |
| chr6_-_112081113 | 0.23 |
ENST00000517419.1
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr3_+_52720187 | 0.23 |
ENST00000474423.1
|
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr11_-_105010320 | 0.23 |
ENST00000532895.1
ENST00000530950.1 |
CARD18
|
caspase recruitment domain family, member 18 |
| chrX_-_152939252 | 0.23 |
ENST00000340888.3
|
PNCK
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr2_+_220144168 | 0.23 |
ENST00000392087.2
ENST00000442681.1 ENST00000439026.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr7_+_2393714 | 0.23 |
ENST00000431643.1
|
EIF3B
|
eukaryotic translation initiation factor 3, subunit B |
| chr22_+_41253080 | 0.23 |
ENST00000541156.1
ENST00000414396.1 ENST00000357137.4 |
XPNPEP3
|
X-prolyl aminopeptidase (aminopeptidase P) 3, putative |
| chr7_+_116451100 | 0.22 |
ENST00000464223.1
ENST00000484325.1 |
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr6_-_116601044 | 0.22 |
ENST00000368608.3
|
TSPYL1
|
TSPY-like 1 |
| chr9_+_19408999 | 0.22 |
ENST00000340967.2
|
ACER2
|
alkaline ceramidase 2 |
| chr4_-_159644507 | 0.22 |
ENST00000307720.3
|
PPID
|
peptidylprolyl isomerase D |
| chr12_-_76425368 | 0.22 |
ENST00000602540.1
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1 |
| chr6_-_38607673 | 0.22 |
ENST00000481247.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr17_+_8339189 | 0.22 |
ENST00000585098.1
ENST00000380025.4 ENST00000402554.3 ENST00000584866.1 ENST00000582490.1 |
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr6_-_131949200 | 0.22 |
ENST00000539158.1
ENST00000368058.1 |
MED23
|
mediator complex subunit 23 |
| chr2_+_231577532 | 0.22 |
ENST00000258418.5
|
CAB39
|
calcium binding protein 39 |
| chr19_+_3762703 | 0.22 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr13_-_79980315 | 0.22 |
ENST00000438737.2
|
RBM26
|
RNA binding motif protein 26 |
| chr1_-_10856694 | 0.22 |
ENST00000377022.3
ENST00000344008.5 |
CASZ1
|
castor zinc finger 1 |
| chr3_+_32433363 | 0.22 |
ENST00000465248.1
|
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr6_-_160210604 | 0.21 |
ENST00000420894.2
ENST00000539756.1 ENST00000544255.1 |
TCP1
|
t-complex 1 |
| chr1_-_149982624 | 0.21 |
ENST00000417191.1
ENST00000369135.4 |
OTUD7B
|
OTU domain containing 7B |
| chr7_+_70229899 | 0.21 |
ENST00000443672.1
|
AUTS2
|
autism susceptibility candidate 2 |
| chr3_-_183735651 | 0.21 |
ENST00000427120.2
ENST00000392579.2 ENST00000382494.2 ENST00000446941.2 |
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr1_-_201390846 | 0.21 |
ENST00000367312.1
ENST00000555340.2 ENST00000361379.4 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr4_+_90032651 | 0.21 |
ENST00000603357.1
|
RP11-84C13.1
|
RP11-84C13.1 |
| chr5_-_171615315 | 0.21 |
ENST00000176763.5
|
STK10
|
serine/threonine kinase 10 |
| chr6_+_32936353 | 0.21 |
ENST00000374825.4
|
BRD2
|
bromodomain containing 2 |
| chr11_-_33744487 | 0.21 |
ENST00000426650.2
|
CD59
|
CD59 molecule, complement regulatory protein |
| chrX_-_152989499 | 0.21 |
ENST00000423827.1
|
BCAP31
|
B-cell receptor-associated protein 31 |
| chr17_-_39928501 | 0.21 |
ENST00000420370.1
|
JUP
|
junction plakoglobin |
| chr2_+_85360499 | 0.21 |
ENST00000282111.3
|
TCF7L1
|
transcription factor 7-like 1 (T-cell specific, HMG-box) |
| chr1_-_68698197 | 0.21 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr22_+_38453207 | 0.21 |
ENST00000404072.3
ENST00000424694.1 |
PICK1
|
protein interacting with PRKCA 1 |
| chr5_+_14664762 | 0.21 |
ENST00000284274.4
|
FAM105B
|
family with sequence similarity 105, member B |
| chr1_+_47489240 | 0.20 |
ENST00000371901.3
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1 |
| chr2_+_113816685 | 0.20 |
ENST00000393200.2
|
IL36RN
|
interleukin 36 receptor antagonist |
| chr3_-_195603566 | 0.20 |
ENST00000424563.1
ENST00000411741.1 |
TNK2
|
tyrosine kinase, non-receptor, 2 |
| chrX_+_64887512 | 0.20 |
ENST00000360270.5
|
MSN
|
moesin |
| chr17_-_76123101 | 0.20 |
ENST00000392467.3
|
TMC6
|
transmembrane channel-like 6 |
| chr9_+_123970052 | 0.20 |
ENST00000373823.3
|
GSN
|
gelsolin |
| chr21_-_46221684 | 0.20 |
ENST00000330942.5
|
UBE2G2
|
ubiquitin-conjugating enzyme E2G 2 |
| chr22_+_29664241 | 0.20 |
ENST00000436425.1
ENST00000447973.1 |
EWSR1
|
EWS RNA-binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HSF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.2 | 1.0 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.2 | 0.6 | GO:0060545 | positive regulation of necroptotic process(GO:0060545) |
| 0.2 | 1.4 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.2 | 0.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.2 | 0.6 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.2 | 1.1 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.2 | 0.5 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.2 | 0.5 | GO:0048633 | negative regulation of auditory receptor cell differentiation(GO:0045608) positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.2 | 0.5 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.1 | 0.7 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 2.5 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.1 | 1.2 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.1 | 0.4 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.1 | 0.6 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.1 | 0.5 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.1 | 0.8 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.3 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.3 | GO:0008052 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.1 | 0.4 | GO:0042000 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.1 | 0.3 | GO:0048003 | synaptic vesicle recycling via endosome(GO:0036466) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 3.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.4 | GO:1904000 | positive regulation of eating behavior(GO:1904000) |
| 0.1 | 0.4 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.1 | 0.6 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.1 | 0.6 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 0.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.4 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.1 | 1.2 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.5 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.2 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.6 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.3 | GO:0021557 | optic cup structural organization(GO:0003409) oculomotor nerve development(GO:0021557) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.1 | 0.2 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.1 | 0.4 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.2 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.4 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 0.6 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.4 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.2 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.1 | 0.4 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 0.3 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 0.7 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.4 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.2 | GO:0086054 | atrial ventricular junction remodeling(GO:0003294) atrial cardiac muscle cell to AV node cell communication by electrical coupling(GO:0086044) bundle of His cell to Purkinje myocyte communication by electrical coupling(GO:0086054) Purkinje myocyte to ventricular cardiac muscle cell communication by electrical coupling(GO:0086055) regulation of Purkinje myocyte action potential(GO:0098906) vasomotion(GO:1990029) |
| 0.1 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.3 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 | 0.2 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.1 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.9 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.1 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.0 | 0.2 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.4 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0032730 | positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.3 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.2 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.7 | GO:0070970 | interleukin-2 secretion(GO:0070970) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.9 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0051466 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.2 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.4 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.2 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 1.2 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.0 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.0 | GO:0002886 | regulation of myeloid leukocyte mediated immunity(GO:0002886) |
| 0.0 | 0.2 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.3 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.3 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.2 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.1 | GO:0019082 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.2 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.2 | GO:0032907 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.0 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.3 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.2 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.0 | 0.8 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.3 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.3 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) negative regulation of hydrogen peroxide catabolic process(GO:2000296) regulation of oxygen metabolic process(GO:2000374) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.3 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.1 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.4 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.0 | GO:1904349 | regulation of small intestine smooth muscle contraction(GO:1904347) positive regulation of small intestine smooth muscle contraction(GO:1904349) small intestine smooth muscle contraction(GO:1990770) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.1 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.2 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:1901907 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.2 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.2 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.3 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.2 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.1 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.0 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.2 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.7 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.0 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 1.0 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.3 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.4 | GO:0043373 | T-helper cell lineage commitment(GO:0002295) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 0.8 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.0 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.0 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.1 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.0 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.4 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.4 | GO:0007620 | copulation(GO:0007620) |
| 0.0 | 0.1 | GO:0051413 | response to cortisone(GO:0051413) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.5 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.1 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.0 | 0.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.4 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.0 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 1.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.3 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0071665 | gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.2 | 2.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.2 | 1.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.7 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 1.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.6 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.1 | 0.4 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 4.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 1.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.5 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.6 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.4 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.3 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 2.1 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.0 | GO:0001534 | radial spoke(GO:0001534) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.2 | 1.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 0.7 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 0.4 | GO:0005365 | myo-inositol transmembrane transporter activity(GO:0005365) |
| 0.1 | 0.6 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.6 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 2.9 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.3 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.1 | 0.5 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.1 | 1.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.2 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 0.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 1.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.2 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.1 | 0.6 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.2 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.1 | 0.9 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.2 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.1 | 0.6 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.9 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 2.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.3 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.3 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.2 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.4 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.2 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.1 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.2 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 3.7 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.5 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.3 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.2 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.1 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.0 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.2 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 3.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.0 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 1.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.0 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.0 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.0 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.0 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 2.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 1.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 1.0 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.6 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 1.5 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |