Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
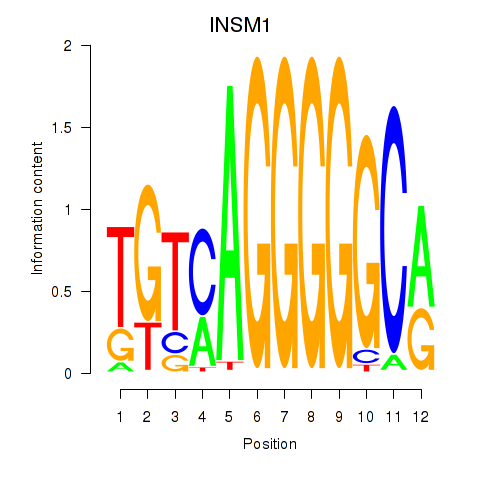
Results for INSM1
Z-value: 0.82
Transcription factors associated with INSM1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
INSM1
|
ENSG00000173404.3 | INSM transcriptional repressor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| INSM1 | hg19_v2_chr20_+_20348740_20348765 | 0.26 | 2.7e-01 | Click! |
Activity profile of INSM1 motif
Sorted Z-values of INSM1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_52887034 | 5.55 |
ENST00000330722.6
|
KRT6A
|
keratin 6A |
| chr3_+_50192499 | 4.07 |
ENST00000413852.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr3_+_50192537 | 3.91 |
ENST00000002829.3
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr3_+_50192457 | 3.82 |
ENST00000414301.1
ENST00000450338.1 |
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr3_+_50192833 | 2.51 |
ENST00000426511.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr1_-_6321035 | 1.52 |
ENST00000377893.2
|
GPR153
|
G protein-coupled receptor 153 |
| chr1_+_160370344 | 1.46 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr10_+_91061712 | 1.44 |
ENST00000371826.3
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr12_-_54779511 | 1.43 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr20_-_45980621 | 1.36 |
ENST00000446894.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr12_+_70760056 | 1.36 |
ENST00000258111.4
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr19_+_10197463 | 1.32 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr11_+_8704748 | 1.28 |
ENST00000526562.1
ENST00000525981.1 |
RPL27A
|
ribosomal protein L27a |
| chr1_+_2398876 | 1.26 |
ENST00000449969.1
|
PLCH2
|
phospholipase C, eta 2 |
| chr14_+_85996507 | 1.14 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr4_+_26322409 | 1.12 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr11_+_124933191 | 1.11 |
ENST00000532000.1
ENST00000308074.4 |
SLC37A2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr19_+_10196981 | 1.10 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr14_+_85996471 | 1.07 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr3_+_10857885 | 1.06 |
ENST00000254488.2
ENST00000454147.1 |
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter), member 11 |
| chr11_+_124932986 | 1.04 |
ENST00000407458.1
ENST00000298280.5 |
SLC37A2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr15_+_45722727 | 0.96 |
ENST00000396650.2
ENST00000558435.1 ENST00000344300.3 |
C15orf48
|
chromosome 15 open reading frame 48 |
| chr3_-_69062764 | 0.94 |
ENST00000295571.5
|
EOGT
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr11_+_124932955 | 0.94 |
ENST00000403796.2
|
SLC37A2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr22_-_20104700 | 0.89 |
ENST00000439169.2
ENST00000445045.1 ENST00000404751.3 ENST00000252136.7 ENST00000403707.3 |
TRMT2A
|
tRNA methyltransferase 2 homolog A (S. cerevisiae) |
| chr4_-_143767428 | 0.89 |
ENST00000513000.1
ENST00000509777.1 ENST00000503927.1 |
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr8_-_57123815 | 0.86 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr18_+_34409069 | 0.85 |
ENST00000543923.1
ENST00000280020.5 ENST00000435985.2 ENST00000592521.1 ENST00000587139.1 |
KIAA1328
|
KIAA1328 |
| chr20_-_45981138 | 0.82 |
ENST00000446994.2
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr1_-_6557156 | 0.82 |
ENST00000537245.1
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr3_-_69062790 | 0.80 |
ENST00000540955.1
ENST00000456376.1 |
EOGT
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr19_+_10196781 | 0.80 |
ENST00000253110.11
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr8_-_11058847 | 0.79 |
ENST00000297303.4
ENST00000416569.2 |
XKR6
|
XK, Kell blood group complex subunit-related family, member 6 |
| chr1_-_154531095 | 0.79 |
ENST00000292211.4
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr10_+_115999089 | 0.77 |
ENST00000392982.3
|
VWA2
|
von Willebrand factor A domain containing 2 |
| chr11_+_8704298 | 0.75 |
ENST00000531978.1
ENST00000524496.1 ENST00000532359.1 ENST00000530022.1 |
RPL27A
|
ribosomal protein L27a |
| chr22_+_20104947 | 0.74 |
ENST00000402752.1
|
RANBP1
|
RAN binding protein 1 |
| chr19_+_19626531 | 0.72 |
ENST00000507754.4
|
NDUFA13
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr3_+_185304059 | 0.71 |
ENST00000427465.2
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr22_+_20105012 | 0.70 |
ENST00000331821.3
ENST00000411892.1 |
RANBP1
|
RAN binding protein 1 |
| chr17_-_1508379 | 0.69 |
ENST00000412517.3
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr1_-_154943002 | 0.68 |
ENST00000606391.1
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr4_-_120548779 | 0.68 |
ENST00000264805.5
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific |
| chr17_+_42081914 | 0.68 |
ENST00000293404.3
ENST00000589767.1 |
NAGS
|
N-acetylglutamate synthase |
| chr21_-_46707793 | 0.67 |
ENST00000331343.7
ENST00000349485.5 |
POFUT2
|
protein O-fucosyltransferase 2 |
| chr5_-_175964366 | 0.64 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr21_-_46238034 | 0.64 |
ENST00000332859.6
|
SUMO3
|
small ubiquitin-like modifier 3 |
| chr15_+_89182178 | 0.62 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr14_-_104313824 | 0.59 |
ENST00000553739.1
ENST00000202556.9 |
PPP1R13B
|
protein phosphatase 1, regulatory subunit 13B |
| chr14_-_85996332 | 0.59 |
ENST00000380722.1
|
RP11-497E19.1
|
RP11-497E19.1 |
| chr11_-_65429891 | 0.59 |
ENST00000527874.1
|
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr22_+_20105259 | 0.58 |
ENST00000416427.1
ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1
|
RAN binding protein 1 |
| chr6_-_41863098 | 0.58 |
ENST00000373006.1
|
USP49
|
ubiquitin specific peptidase 49 |
| chr11_-_65430251 | 0.58 |
ENST00000534283.1
ENST00000527749.1 ENST00000533187.1 ENST00000525693.1 ENST00000534558.1 ENST00000532879.1 ENST00000532999.1 |
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr3_-_69062742 | 0.58 |
ENST00000424374.1
|
EOGT
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr1_+_65613217 | 0.57 |
ENST00000545314.1
|
AK4
|
adenylate kinase 4 |
| chr15_+_89181974 | 0.56 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr10_+_115999004 | 0.56 |
ENST00000603594.1
|
VWA2
|
von Willebrand factor A domain containing 2 |
| chr1_+_65613340 | 0.56 |
ENST00000546702.1
|
AK4
|
adenylate kinase 4 |
| chr17_-_7297833 | 0.55 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr2_+_24272576 | 0.55 |
ENST00000380986.4
ENST00000452109.1 |
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa |
| chr13_-_52027134 | 0.55 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr15_-_43802769 | 0.54 |
ENST00000263801.3
|
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr18_-_34409116 | 0.54 |
ENST00000334295.4
|
TPGS2
|
tubulin polyglutamylase complex subunit 2 |
| chr18_-_34408902 | 0.54 |
ENST00000593035.1
ENST00000383056.3 ENST00000588909.1 ENST00000590337.1 |
TPGS2
|
tubulin polyglutamylase complex subunit 2 |
| chr6_+_155054459 | 0.53 |
ENST00000367178.3
ENST00000417268.1 ENST00000367186.4 |
SCAF8
|
SR-related CTD-associated factor 8 |
| chr9_-_112083229 | 0.52 |
ENST00000374566.3
ENST00000374557.4 |
EPB41L4B
|
erythrocyte membrane protein band 4.1 like 4B |
| chr9_-_140115775 | 0.52 |
ENST00000391553.1
ENST00000392827.1 |
RNF208
|
ring finger protein 208 |
| chr21_-_46237959 | 0.51 |
ENST00000397898.3
ENST00000411651.2 |
SUMO3
|
small ubiquitin-like modifier 3 |
| chr17_+_45608430 | 0.50 |
ENST00000322157.4
|
NPEPPS
|
aminopeptidase puromycin sensitive |
| chr14_+_102027688 | 0.49 |
ENST00000510508.4
ENST00000359323.3 |
DIO3
|
deiodinase, iodothyronine, type III |
| chr8_-_116681686 | 0.48 |
ENST00000519815.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chrX_+_146993648 | 0.48 |
ENST00000370470.1
|
FMR1
|
fragile X mental retardation 1 |
| chr4_-_151936865 | 0.48 |
ENST00000535741.1
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr11_-_57092381 | 0.48 |
ENST00000358252.3
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chrX_+_69674943 | 0.47 |
ENST00000542398.1
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr3_+_185303962 | 0.46 |
ENST00000296257.5
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr8_-_124286495 | 0.46 |
ENST00000297857.2
|
ZHX1
|
zinc fingers and homeoboxes 1 |
| chr15_+_89182156 | 0.46 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr13_-_52026730 | 0.45 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr15_+_79166065 | 0.44 |
ENST00000559690.1
ENST00000559158.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr3_-_48601206 | 0.44 |
ENST00000273610.3
|
UCN2
|
urocortin 2 |
| chr3_+_14989186 | 0.43 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr11_-_65430554 | 0.43 |
ENST00000308639.9
ENST00000406246.3 |
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr2_+_232575128 | 0.42 |
ENST00000412128.1
|
PTMA
|
prothymosin, alpha |
| chr2_+_24272543 | 0.42 |
ENST00000380991.4
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa |
| chr16_+_610407 | 0.42 |
ENST00000409413.3
|
C16orf11
|
chromosome 16 open reading frame 11 |
| chr4_-_129209221 | 0.41 |
ENST00000512483.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr4_-_151936416 | 0.41 |
ENST00000510413.1
ENST00000507224.1 |
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr1_-_154943212 | 0.40 |
ENST00000368445.5
ENST00000448116.2 ENST00000368449.4 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr2_-_153573887 | 0.40 |
ENST00000493468.2
ENST00000545856.1 |
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr5_-_11904152 | 0.39 |
ENST00000304623.8
ENST00000458100.2 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr11_-_46142615 | 0.38 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr12_-_80328700 | 0.37 |
ENST00000550107.1
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A |
| chr6_+_32938665 | 0.37 |
ENST00000374831.4
ENST00000395289.2 |
BRD2
|
bromodomain containing 2 |
| chr11_+_118307179 | 0.36 |
ENST00000534358.1
ENST00000531904.2 ENST00000389506.5 ENST00000354520.4 |
KMT2A
|
lysine (K)-specific methyltransferase 2A |
| chr15_-_70388943 | 0.36 |
ENST00000559048.1
ENST00000560939.1 ENST00000440567.3 ENST00000557907.1 ENST00000558379.1 ENST00000451782.2 ENST00000559929.1 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr11_+_120207787 | 0.36 |
ENST00000397843.2
ENST00000356641.3 |
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr2_-_74667612 | 0.35 |
ENST00000305557.5
ENST00000233330.6 |
RTKN
|
rhotekin |
| chr16_-_30134441 | 0.34 |
ENST00000395200.1
|
MAPK3
|
mitogen-activated protein kinase 3 |
| chr6_+_32938692 | 0.33 |
ENST00000443797.2
|
BRD2
|
bromodomain containing 2 |
| chr1_-_39325431 | 0.33 |
ENST00000373001.3
|
RRAGC
|
Ras-related GTP binding C |
| chr5_-_11904100 | 0.32 |
ENST00000359640.2
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr5_-_141030943 | 0.32 |
ENST00000522783.1
ENST00000519800.1 ENST00000435817.2 |
FCHSD1
|
FCH and double SH3 domains 1 |
| chr7_-_91509986 | 0.32 |
ENST00000456229.1
ENST00000442961.1 ENST00000406735.2 ENST00000419292.1 ENST00000351870.3 |
MTERF
|
mitochondrial transcription termination factor |
| chr10_-_104001231 | 0.31 |
ENST00000370002.3
|
PITX3
|
paired-like homeodomain 3 |
| chr22_+_47158578 | 0.31 |
ENST00000355704.3
|
TBC1D22A
|
TBC1 domain family, member 22A |
| chr17_-_7297519 | 0.31 |
ENST00000576362.1
ENST00000571078.1 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr8_-_120685608 | 0.31 |
ENST00000427067.2
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr7_-_91509972 | 0.30 |
ENST00000425936.1
|
MTERF
|
mitochondrial transcription termination factor |
| chr17_-_32906388 | 0.30 |
ENST00000357754.1
|
C17orf102
|
chromosome 17 open reading frame 102 |
| chr12_+_3000037 | 0.30 |
ENST00000544943.1
ENST00000448120.2 |
TULP3
|
tubby like protein 3 |
| chr9_-_34637718 | 0.30 |
ENST00000378892.1
ENST00000277010.4 |
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr6_-_29595779 | 0.29 |
ENST00000355973.3
ENST00000377012.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chrX_-_71525742 | 0.29 |
ENST00000450875.1
ENST00000417400.1 ENST00000431381.1 ENST00000445983.1 |
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr6_+_108881012 | 0.29 |
ENST00000343882.6
|
FOXO3
|
forkhead box O3 |
| chr17_-_4890919 | 0.29 |
ENST00000572543.1
ENST00000381311.5 ENST00000348066.3 ENST00000358183.4 |
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr15_+_43803143 | 0.28 |
ENST00000382031.1
|
MAP1A
|
microtubule-associated protein 1A |
| chrX_+_37430822 | 0.27 |
ENST00000378621.3
ENST00000378619.3 |
LANCL3
|
LanC lantibiotic synthetase component C-like 3 (bacterial) |
| chr1_-_37980344 | 0.26 |
ENST00000448519.2
ENST00000373075.2 ENST00000373073.4 ENST00000296214.5 |
MEAF6
|
MYST/Esa1-associated factor 6 |
| chr6_-_38607673 | 0.26 |
ENST00000481247.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr6_+_108882069 | 0.26 |
ENST00000406360.1
|
FOXO3
|
forkhead box O3 |
| chr16_+_88704978 | 0.26 |
ENST00000244241.4
|
IL17C
|
interleukin 17C |
| chr18_-_34408802 | 0.26 |
ENST00000590842.1
|
TPGS2
|
tubulin polyglutamylase complex subunit 2 |
| chr12_+_58148842 | 0.25 |
ENST00000266643.5
|
MARCH9
|
membrane-associated ring finger (C3HC4) 9 |
| chr7_-_132262060 | 0.25 |
ENST00000359827.3
|
PLXNA4
|
plexin A4 |
| chr11_+_117049445 | 0.24 |
ENST00000324225.4
ENST00000532960.1 |
SIDT2
|
SID1 transmembrane family, member 2 |
| chrX_+_146993534 | 0.24 |
ENST00000334557.6
ENST00000439526.2 ENST00000370475.4 |
FMR1
|
fragile X mental retardation 1 |
| chr19_+_48972459 | 0.23 |
ENST00000427476.1
|
CYTH2
|
cytohesin 2 |
| chr1_+_230203010 | 0.23 |
ENST00000541865.1
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr4_-_147443043 | 0.23 |
ENST00000394059.4
ENST00000502607.1 ENST00000335472.7 ENST00000432059.2 ENST00000394062.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr14_+_62162258 | 0.22 |
ENST00000337138.4
ENST00000394997.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chrX_-_153059958 | 0.22 |
ENST00000370092.3
ENST00000217901.5 |
IDH3G
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr1_+_154474689 | 0.22 |
ENST00000368482.4
|
TDRD10
|
tudor domain containing 10 |
| chr22_+_44351419 | 0.22 |
ENST00000396202.3
|
SAMM50
|
SAMM50 sorting and assembly machinery component |
| chr14_-_62162541 | 0.22 |
ENST00000557544.1
|
HIF1A-AS1
|
HIF1A antisense RNA 1 |
| chrX_-_153059811 | 0.22 |
ENST00000427365.2
ENST00000444450.1 ENST00000370093.1 |
IDH3G
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr12_-_118810688 | 0.22 |
ENST00000542532.1
ENST00000392533.3 |
TAOK3
|
TAO kinase 3 |
| chr8_+_145065705 | 0.21 |
ENST00000533044.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr14_-_77227695 | 0.21 |
ENST00000556271.1
|
RP11-99E15.2
|
RP11-99E15.2 |
| chr11_+_10772534 | 0.21 |
ENST00000361367.2
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr17_+_7211280 | 0.20 |
ENST00000419711.2
ENST00000571955.1 ENST00000573714.1 |
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr12_-_76953513 | 0.20 |
ENST00000547540.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr4_+_619386 | 0.20 |
ENST00000496514.1
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr8_-_145550337 | 0.20 |
ENST00000531896.1
|
DGAT1
|
diacylglycerol O-acyltransferase 1 |
| chr7_+_100318423 | 0.19 |
ENST00000252723.2
|
EPO
|
erythropoietin |
| chr4_+_619347 | 0.18 |
ENST00000255622.6
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr15_-_74045088 | 0.18 |
ENST00000569673.1
|
C15orf59
|
chromosome 15 open reading frame 59 |
| chr6_-_17706618 | 0.18 |
ENST00000262077.2
ENST00000537253.1 |
NUP153
|
nucleoporin 153kDa |
| chr1_+_53098862 | 0.18 |
ENST00000517870.1
|
FAM159A
|
family with sequence similarity 159, member A |
| chr20_+_44657807 | 0.17 |
ENST00000372315.1
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr20_+_35202909 | 0.17 |
ENST00000558530.1
ENST00000558028.1 ENST00000560025.1 |
TGIF2-C20orf24
TGIF2
|
TGIF2-C20orf24 readthrough TGFB-induced factor homeobox 2 |
| chr9_+_136243264 | 0.17 |
ENST00000371955.1
|
C9orf96
|
chromosome 9 open reading frame 96 |
| chr1_-_85725316 | 0.17 |
ENST00000344356.5
ENST00000471115.1 |
C1orf52
|
chromosome 1 open reading frame 52 |
| chr1_-_36851475 | 0.17 |
ENST00000373129.3
|
STK40
|
serine/threonine kinase 40 |
| chr15_-_96590126 | 0.17 |
ENST00000561051.1
|
RP11-4G2.1
|
RP11-4G2.1 |
| chr11_+_120110863 | 0.16 |
ENST00000543440.2
|
POU2F3
|
POU class 2 homeobox 3 |
| chrX_+_153059608 | 0.16 |
ENST00000370087.1
|
SSR4
|
signal sequence receptor, delta |
| chr18_+_46065570 | 0.16 |
ENST00000591412.1
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr9_-_130517522 | 0.16 |
ENST00000373274.3
ENST00000420366.1 |
SH2D3C
|
SH2 domain containing 3C |
| chr17_-_37382105 | 0.16 |
ENST00000333461.5
|
STAC2
|
SH3 and cysteine rich domain 2 |
| chr1_-_99470368 | 0.16 |
ENST00000263177.4
|
LPPR5
|
Lipid phosphate phosphatase-related protein type 5 |
| chr22_+_47158518 | 0.15 |
ENST00000337137.4
ENST00000380995.1 ENST00000407381.3 |
TBC1D22A
|
TBC1 domain family, member 22A |
| chr16_+_23569021 | 0.15 |
ENST00000567212.1
ENST00000567264.1 |
UBFD1
|
ubiquitin family domain containing 1 |
| chr18_+_46065483 | 0.15 |
ENST00000382998.4
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr12_-_76953453 | 0.15 |
ENST00000549570.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr13_-_36871886 | 0.14 |
ENST00000491049.2
ENST00000503173.1 ENST00000239860.6 ENST00000379862.2 ENST00000239859.7 ENST00000379864.2 ENST00000510088.1 ENST00000554962.1 ENST00000511166.1 |
CCDC169
SOHLH2
CCDC169-SOHLH2
|
coiled-coil domain containing 169 spermatogenesis and oogenesis specific basic helix-loop-helix 2 CCDC169-SOHLH2 readthrough |
| chr16_-_56553882 | 0.14 |
ENST00000568104.1
|
BBS2
|
Bardet-Biedl syndrome 2 |
| chr16_-_30134524 | 0.14 |
ENST00000395202.1
ENST00000395199.3 ENST00000263025.4 ENST00000322266.5 ENST00000403394.1 |
MAPK3
|
mitogen-activated protein kinase 3 |
| chrX_-_41782249 | 0.14 |
ENST00000442742.2
ENST00000421587.2 ENST00000378166.4 ENST00000318588.9 ENST00000361962.4 |
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr1_+_156863470 | 0.14 |
ENST00000338302.3
ENST00000455314.1 ENST00000292357.7 |
PEAR1
|
platelet endothelial aggregation receptor 1 |
| chr2_-_159313214 | 0.14 |
ENST00000409889.1
ENST00000283233.5 ENST00000536771.1 |
CCDC148
|
coiled-coil domain containing 148 |
| chr1_-_154474589 | 0.14 |
ENST00000304760.2
|
SHE
|
Src homology 2 domain containing E |
| chr19_-_48894104 | 0.13 |
ENST00000597017.1
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr1_-_85666688 | 0.13 |
ENST00000341460.5
|
SYDE2
|
synapse defective 1, Rho GTPase, homolog 2 (C. elegans) |
| chr15_-_52263937 | 0.12 |
ENST00000315141.5
ENST00000299601.5 |
LEO1
|
Leo1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) |
| chr14_+_106938440 | 0.12 |
ENST00000433371.1
ENST00000449670.1 ENST00000334298.3 |
LINC00221
|
long intergenic non-protein coding RNA 221 |
| chr17_-_46623441 | 0.12 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr17_-_56606664 | 0.12 |
ENST00000580844.1
|
SEPT4
|
septin 4 |
| chr9_-_37034028 | 0.12 |
ENST00000520281.1
ENST00000446742.1 ENST00000522003.1 ENST00000523145.1 ENST00000414447.1 ENST00000377847.2 ENST00000377853.2 ENST00000377852.2 ENST00000523241.1 ENST00000520154.1 ENST00000358127.4 |
PAX5
|
paired box 5 |
| chr6_-_31125850 | 0.12 |
ENST00000507751.1
ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1
|
coiled-coil alpha-helical rod protein 1 |
| chr15_+_27216530 | 0.12 |
ENST00000555083.1
|
GABRG3
|
gamma-aminobutyric acid (GABA) A receptor, gamma 3 |
| chr14_-_51297197 | 0.11 |
ENST00000382043.4
|
NIN
|
ninein (GSK3B interacting protein) |
| chr17_-_56606705 | 0.11 |
ENST00000317268.3
|
SEPT4
|
septin 4 |
| chr1_-_36851489 | 0.11 |
ENST00000373130.3
ENST00000373132.3 |
STK40
|
serine/threonine kinase 40 |
| chr10_+_76585303 | 0.11 |
ENST00000372725.1
|
KAT6B
|
K(lysine) acetyltransferase 6B |
| chr12_+_120105558 | 0.10 |
ENST00000229328.5
ENST00000541640.1 |
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr11_-_46142948 | 0.10 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chr4_+_56815102 | 0.10 |
ENST00000257287.4
|
CEP135
|
centrosomal protein 135kDa |
| chr7_-_15726296 | 0.10 |
ENST00000262041.5
|
MEOX2
|
mesenchyme homeobox 2 |
| chr17_-_56606639 | 0.10 |
ENST00000579371.1
|
SEPT4
|
septin 4 |
| chr18_-_51750948 | 0.09 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr18_+_46065393 | 0.09 |
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr4_-_129208940 | 0.09 |
ENST00000296425.5
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr1_+_168148273 | 0.09 |
ENST00000367830.3
|
TIPRL
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr22_-_30695471 | 0.09 |
ENST00000434291.1
|
RP1-130H16.18
|
Uncharacterized protein |
| chr16_+_67360712 | 0.09 |
ENST00000569499.2
ENST00000329956.6 ENST00000561948.1 |
LRRC36
|
leucine rich repeat containing 36 |
| chr20_+_44657845 | 0.09 |
ENST00000243964.3
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr2_+_232575168 | 0.09 |
ENST00000440384.1
|
PTMA
|
prothymosin, alpha |
| chr5_+_178986693 | 0.09 |
ENST00000437570.2
ENST00000393438.2 |
RUFY1
|
RUN and FYVE domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of INSM1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.6 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 1.0 | 14.3 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.5 | 1.6 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.5 | 1.5 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.4 | 2.2 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.3 | 1.6 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 | 3.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.3 | 0.8 | GO:1901254 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.3 | 2.0 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.2 | 2.2 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.2 | 1.4 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 0.5 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.2 | 0.2 | GO:0046832 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.2 | 0.3 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.2 | 0.5 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.1 | 0.6 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 1.1 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.4 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.1 | 1.1 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.1 | 1.3 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 | 1.4 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 0.3 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.1 | 0.7 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.2 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.1 | 1.0 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 1.2 | GO:0035562 | protein desumoylation(GO:0016926) negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 0.7 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.6 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.7 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.3 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 0.2 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.1 | 0.4 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.3 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.1 | 1.1 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 | 0.5 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.1 | 0.8 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 0.2 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.1 | 0.4 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.5 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 0.3 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 1.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.7 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.9 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.5 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.3 | GO:0071105 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.0 | 1.0 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.3 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.3 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.1 | GO:2001162 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.0 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.5 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 1.1 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 1.2 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 0.3 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 2.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.8 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 2.1 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 1.0 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 2.2 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.5 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.9 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.0 | 0.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.4 | 1.5 | GO:0060187 | cell pole(GO:0060187) |
| 0.3 | 0.8 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.1 | 1.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 1.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.3 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 4.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.3 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 2.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 4.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 2.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.3 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 0.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.5 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 16.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.5 | 1.6 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.4 | 2.3 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.4 | 3.1 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 1.0 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.2 | 0.2 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.2 | 2.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 1.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.9 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 1.2 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.7 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 1.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 1.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.4 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.8 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.4 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.4 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 1.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.5 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.3 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 2.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 2.0 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 1.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.3 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 1.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 1.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 1.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 5.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.3 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 1.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.9 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 14.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.8 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 2.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 1.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 3.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 2.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 2.2 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |