Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
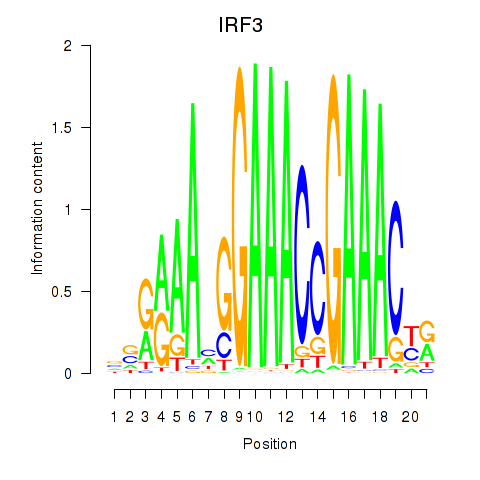
Results for IRF3
Z-value: 1.54
Transcription factors associated with IRF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRF3
|
ENSG00000126456.11 | interferon regulatory factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF3 | hg19_v2_chr19_-_50169064_50169132 | 0.57 | 9.1e-03 | Click! |
Activity profile of IRF3 motif
Sorted Z-values of IRF3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_948803 | 7.84 |
ENST00000379389.4
|
ISG15
|
ISG15 ubiquitin-like modifier |
| chr10_+_91087651 | 5.62 |
ENST00000371818.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr1_+_79115503 | 4.96 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr10_+_91092241 | 4.76 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr21_+_42792442 | 4.13 |
ENST00000398600.2
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr2_-_163175133 | 4.06 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr12_+_113416191 | 3.77 |
ENST00000342315.4
ENST00000392583.2 |
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr3_-_172241250 | 3.64 |
ENST00000420541.2
ENST00000241261.2 |
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr11_-_615570 | 3.48 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr11_-_615942 | 3.44 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr22_+_36649056 | 3.36 |
ENST00000397278.3
ENST00000422706.1 ENST00000426053.1 ENST00000319136.4 |
APOL1
|
apolipoprotein L, 1 |
| chr12_+_113416265 | 3.30 |
ENST00000449768.2
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr19_-_17516449 | 3.27 |
ENST00000252593.6
|
BST2
|
bone marrow stromal cell antigen 2 |
| chr10_+_91061712 | 2.90 |
ENST00000371826.3
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr22_+_36649170 | 2.65 |
ENST00000438034.1
ENST00000427990.1 ENST00000347595.7 ENST00000397279.4 ENST00000433768.1 ENST00000440669.2 |
APOL1
|
apolipoprotein L, 1 |
| chr7_-_92747269 | 2.60 |
ENST00000446617.1
ENST00000379958.2 |
SAMD9
|
sterile alpha motif domain containing 9 |
| chr17_+_6659153 | 2.54 |
ENST00000441631.1
ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1
|
XIAP associated factor 1 |
| chr19_+_10196781 | 2.54 |
ENST00000253110.11
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr16_-_11350036 | 2.52 |
ENST00000332029.2
|
SOCS1
|
suppressor of cytokine signaling 1 |
| chr2_-_231084820 | 2.25 |
ENST00000258382.5
ENST00000338556.3 |
SP110
|
SP110 nuclear body protein |
| chr2_-_231084659 | 2.25 |
ENST00000258381.6
ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110
|
SP110 nuclear body protein |
| chr7_-_92777606 | 2.24 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr12_-_10022735 | 2.16 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr19_+_10196981 | 2.13 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr22_-_36635684 | 2.11 |
ENST00000358502.5
|
APOL2
|
apolipoprotein L, 2 |
| chr3_-_122283100 | 2.06 |
ENST00000492382.1
ENST00000462315.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr3_-_122283424 | 1.77 |
ENST00000477522.2
ENST00000360356.2 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr12_+_113416340 | 1.73 |
ENST00000552756.1
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr19_+_39786962 | 1.70 |
ENST00000333625.2
|
IFNL1
|
interferon, lambda 1 |
| chr3_-_122283079 | 1.64 |
ENST00000471785.1
ENST00000466126.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chrX_+_57313113 | 1.57 |
ENST00000374900.4
|
FAAH2
|
fatty acid amide hydrolase 2 |
| chr9_+_5450503 | 1.56 |
ENST00000381573.4
ENST00000381577.3 |
CD274
|
CD274 molecule |
| chr2_-_231084617 | 1.49 |
ENST00000409815.2
|
SP110
|
SP110 nuclear body protein |
| chr11_-_57335280 | 1.48 |
ENST00000287156.4
|
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr7_-_139756791 | 1.47 |
ENST00000489809.1
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr3_+_187086120 | 1.44 |
ENST00000259030.2
|
RTP4
|
receptor (chemosensory) transporter protein 4 |
| chr22_-_36635563 | 1.42 |
ENST00000451256.2
|
APOL2
|
apolipoprotein L, 2 |
| chr1_+_150480551 | 1.32 |
ENST00000369049.4
ENST00000369047.4 |
ECM1
|
extracellular matrix protein 1 |
| chr17_+_25958174 | 1.29 |
ENST00000313648.6
ENST00000577392.1 ENST00000584661.1 ENST00000413914.2 |
LGALS9
|
lectin, galactoside-binding, soluble, 9 |
| chr9_-_32526299 | 1.28 |
ENST00000379882.1
ENST00000379883.2 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr22_-_36635598 | 1.26 |
ENST00000454728.1
|
APOL2
|
apolipoprotein L, 2 |
| chr17_-_7166500 | 1.25 |
ENST00000575313.1
ENST00000397317.4 |
CLDN7
|
claudin 7 |
| chr6_+_32821924 | 1.23 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr1_-_161015752 | 1.22 |
ENST00000435396.1
ENST00000368021.3 |
USF1
|
upstream transcription factor 1 |
| chr2_+_218933972 | 1.18 |
ENST00000374155.3
|
RUFY4
|
RUN and FYVE domain containing 4 |
| chr4_-_169239921 | 1.18 |
ENST00000514995.1
ENST00000393743.3 |
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr11_+_124735282 | 1.17 |
ENST00000397801.1
|
ROBO3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr1_+_159931002 | 1.15 |
ENST00000443364.1
ENST00000423943.1 |
RP11-48O20.4
|
long intergenic non-protein coding RNA 1133 |
| chr7_+_142829162 | 1.13 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr22_-_36556821 | 1.11 |
ENST00000531095.1
ENST00000397293.2 ENST00000349314.2 |
APOL3
|
apolipoprotein L, 3 |
| chr11_-_57334732 | 1.10 |
ENST00000526659.1
ENST00000527022.1 |
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr6_-_36355513 | 1.10 |
ENST00000340181.4
ENST00000373737.4 |
ETV7
|
ets variant 7 |
| chr22_+_36044411 | 1.10 |
ENST00000409652.4
|
APOL6
|
apolipoprotein L, 6 |
| chr4_+_118955500 | 1.07 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr19_+_49977466 | 1.07 |
ENST00000596435.1
ENST00000344019.3 ENST00000597551.1 ENST00000204637.2 ENST00000600429.1 |
FLT3LG
|
fms-related tyrosine kinase 3 ligand |
| chr20_-_47894569 | 1.06 |
ENST00000371744.1
ENST00000371752.1 ENST00000396105.1 |
ZNFX1
|
zinc finger, NFX1-type containing 1 |
| chr5_-_95158375 | 1.05 |
ENST00000512469.2
ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX
|
glutaredoxin (thioltransferase) |
| chr1_+_150480576 | 1.05 |
ENST00000346569.6
|
ECM1
|
extracellular matrix protein 1 |
| chr5_-_95158644 | 1.04 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
| chr22_-_36635225 | 1.00 |
ENST00000529194.1
|
APOL2
|
apolipoprotein L, 2 |
| chr9_-_32526184 | 0.99 |
ENST00000545044.1
ENST00000379868.1 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr1_-_177939041 | 0.98 |
ENST00000308284.6
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr6_-_32821599 | 0.97 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr4_-_156875003 | 0.97 |
ENST00000433477.3
|
CTSO
|
cathepsin O |
| chr12_-_99288536 | 0.96 |
ENST00000549797.1
ENST00000333732.7 ENST00000341752.7 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr22_+_18632666 | 0.91 |
ENST00000215794.7
|
USP18
|
ubiquitin specific peptidase 18 |
| chr11_-_4414880 | 0.89 |
ENST00000254436.7
ENST00000543625.1 |
TRIM21
|
tripartite motif containing 21 |
| chr2_-_73869508 | 0.87 |
ENST00000272425.3
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative) |
| chr8_+_39770803 | 0.86 |
ENST00000518237.1
|
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr6_-_32806506 | 0.84 |
ENST00000374897.2
ENST00000452392.2 |
TAP2
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr11_+_5646213 | 0.83 |
ENST00000429814.2
|
TRIM34
|
tripartite motif containing 34 |
| chr6_+_28227063 | 0.83 |
ENST00000343684.3
|
NKAPL
|
NFKB activating protein-like |
| chr9_-_14322319 | 0.83 |
ENST00000606230.1
|
NFIB
|
nuclear factor I/B |
| chr9_-_100954910 | 0.83 |
ENST00000375077.4
|
CORO2A
|
coronin, actin binding protein, 2A |
| chr6_-_36355486 | 0.81 |
ENST00000538992.1
|
ETV7
|
ets variant 7 |
| chr2_-_166651152 | 0.79 |
ENST00000431484.1
ENST00000412248.1 |
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) |
| chr18_-_8337038 | 0.78 |
ENST00000594251.1
|
AP001094.1
|
Uncharacterized protein |
| chr1_-_161015663 | 0.78 |
ENST00000534633.1
|
USF1
|
upstream transcription factor 1 |
| chr12_+_14369524 | 0.74 |
ENST00000538329.1
|
RP11-134N1.2
|
RP11-134N1.2 |
| chr2_-_166651191 | 0.72 |
ENST00000392701.3
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) |
| chr1_-_89664595 | 0.71 |
ENST00000355754.6
|
GBP4
|
guanylate binding protein 4 |
| chr4_+_130017268 | 0.68 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr3_+_122399444 | 0.67 |
ENST00000474629.2
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr4_+_89378261 | 0.66 |
ENST00000264350.3
|
HERC5
|
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chr17_+_6659354 | 0.65 |
ENST00000574907.1
|
XAF1
|
XIAP associated factor 1 |
| chr19_+_50432400 | 0.64 |
ENST00000423777.2
ENST00000600336.1 ENST00000597227.1 |
ATF5
|
activating transcription factor 5 |
| chr12_-_133613794 | 0.63 |
ENST00000443154.3
|
RP11-386I8.6
|
RP11-386I8.6 |
| chr12_-_133787772 | 0.62 |
ENST00000545350.1
|
AC226150.4
|
Uncharacterized protein |
| chr3_+_122283064 | 0.60 |
ENST00000296161.4
|
DTX3L
|
deltex 3-like (Drosophila) |
| chr2_-_157198860 | 0.60 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr20_+_61436146 | 0.59 |
ENST00000290291.6
|
OGFR
|
opioid growth factor receptor |
| chr20_+_388791 | 0.59 |
ENST00000441733.1
ENST00000353660.3 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr20_+_388679 | 0.58 |
ENST00000356286.5
ENST00000475269.1 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr17_-_40264692 | 0.58 |
ENST00000591220.1
ENST00000251642.3 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr9_+_77112244 | 0.58 |
ENST00000376896.3
|
RORB
|
RAR-related orphan receptor B |
| chr15_-_55611306 | 0.57 |
ENST00000563262.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr7_-_32931387 | 0.56 |
ENST00000304056.4
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr14_-_24615523 | 0.56 |
ENST00000559056.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr19_+_13051206 | 0.54 |
ENST00000586760.1
|
CALR
|
calreticulin |
| chr6_-_32806483 | 0.52 |
ENST00000374899.4
|
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr6_-_3752222 | 0.51 |
ENST00000380283.4
|
PXDC1
|
PX domain containing 1 |
| chr6_+_41748500 | 0.51 |
ENST00000458694.1
ENST00000359201.5 |
PRICKLE4
|
prickle homolog 4 (Drosophila) |
| chr14_-_24615805 | 0.51 |
ENST00000560410.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr10_-_49701686 | 0.50 |
ENST00000417247.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr15_+_45003675 | 0.50 |
ENST00000558401.1
ENST00000559916.1 ENST00000544417.1 |
B2M
|
beta-2-microglobulin |
| chr12_+_133614119 | 0.46 |
ENST00000327668.7
|
ZNF84
|
zinc finger protein 84 |
| chr15_+_64428529 | 0.46 |
ENST00000560861.1
|
SNX1
|
sorting nexin 1 |
| chr12_+_6561190 | 0.45 |
ENST00000544021.1
ENST00000266556.7 |
TAPBPL
|
TAP binding protein-like |
| chr2_-_214017151 | 0.44 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr4_-_169401628 | 0.44 |
ENST00000514748.1
ENST00000512371.1 ENST00000260184.7 ENST00000505890.1 ENST00000511577.1 |
DDX60L
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like |
| chr7_-_138794394 | 0.44 |
ENST00000242351.5
ENST00000471652.1 |
ZC3HAV1
|
zinc finger CCCH-type, antiviral 1 |
| chr1_-_161014731 | 0.43 |
ENST00000368020.1
|
USF1
|
upstream transcription factor 1 |
| chr20_+_388935 | 0.43 |
ENST00000382181.2
ENST00000400247.3 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr12_-_54582655 | 0.43 |
ENST00000504338.1
ENST00000514685.1 ENST00000504797.1 ENST00000513838.1 ENST00000505128.1 ENST00000337581.3 ENST00000503306.1 ENST00000243112.5 ENST00000514196.1 ENST00000506169.1 ENST00000507904.1 ENST00000508394.2 |
SMUG1
|
single-strand-selective monofunctional uracil-DNA glycosylase 1 |
| chr5_-_77656175 | 0.43 |
ENST00000513755.1
ENST00000421004.3 |
CTD-2037K23.2
|
CTD-2037K23.2 |
| chr12_+_133613937 | 0.41 |
ENST00000539354.1
ENST00000542874.1 ENST00000438628.2 |
ZNF84
|
zinc finger protein 84 |
| chr22_+_39436862 | 0.41 |
ENST00000381565.2
ENST00000452957.2 |
APOBEC3F
APOBEC3G
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G |
| chr11_-_67418120 | 0.39 |
ENST00000255082.3
|
ACY3
|
aspartoacylase (aminocyclase) 3 |
| chr12_+_113376157 | 0.39 |
ENST00000228928.7
|
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chrX_+_77154935 | 0.39 |
ENST00000481445.1
|
COX7B
|
cytochrome c oxidase subunit VIIb |
| chr1_-_177939348 | 0.39 |
ENST00000464631.2
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr5_+_131746575 | 0.37 |
ENST00000337752.2
ENST00000378947.3 ENST00000407797.1 |
C5orf56
|
chromosome 5 open reading frame 56 |
| chrX_+_146993648 | 0.37 |
ENST00000370470.1
|
FMR1
|
fragile X mental retardation 1 |
| chrX_+_52920336 | 0.37 |
ENST00000452154.2
|
FAM156B
|
family with sequence similarity 156, member B |
| chr12_+_133613878 | 0.35 |
ENST00000392319.2
ENST00000543758.1 |
ZNF84
|
zinc finger protein 84 |
| chr10_-_120355149 | 0.34 |
ENST00000239032.2
|
PRLHR
|
prolactin releasing hormone receptor |
| chr12_-_4758159 | 0.34 |
ENST00000545990.2
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr14_+_24605361 | 0.34 |
ENST00000206451.6
ENST00000559123.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr17_+_6658878 | 0.33 |
ENST00000574394.1
|
XAF1
|
XIAP associated factor 1 |
| chr22_+_31892261 | 0.32 |
ENST00000432498.1
ENST00000540643.1 ENST00000443326.1 ENST00000414585.1 |
SFI1
|
Sfi1 homolog, spindle assembly associated (yeast) |
| chr6_+_28193037 | 0.32 |
ENST00000531981.1
ENST00000425468.2 ENST00000252207.5 ENST00000531979.1 ENST00000527436.1 |
ZSCAN9
|
zinc finger and SCAN domain containing 9 |
| chr6_-_28226984 | 0.31 |
ENST00000423974.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr1_-_159046617 | 0.30 |
ENST00000368130.4
|
AIM2
|
absent in melanoma 2 |
| chr1_+_110577229 | 0.28 |
ENST00000369795.3
ENST00000369794.2 |
STRIP1
|
striatin interacting protein 1 |
| chr12_-_8218997 | 0.27 |
ENST00000307637.4
|
C3AR1
|
complement component 3a receptor 1 |
| chr6_-_30181133 | 0.26 |
ENST00000454678.2
ENST00000434785.1 |
TRIM26
|
tripartite motif containing 26 |
| chr11_-_110968081 | 0.26 |
ENST00000603154.1
|
RP11-89C3.4
|
RP11-89C3.4 |
| chr17_+_36861735 | 0.25 |
ENST00000378137.5
ENST00000325718.7 |
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr14_-_91976794 | 0.25 |
ENST00000555462.1
|
SMEK1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr14_+_73525144 | 0.25 |
ENST00000261973.7
ENST00000540173.1 |
RBM25
|
RNA binding motif protein 25 |
| chr7_-_122526411 | 0.23 |
ENST00000449022.2
|
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr5_-_180288248 | 0.23 |
ENST00000512132.1
ENST00000506439.1 ENST00000502412.1 ENST00000359141.6 |
ZFP62
|
ZFP62 zinc finger protein |
| chr2_+_127413677 | 0.22 |
ENST00000356887.7
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr16_+_66968343 | 0.21 |
ENST00000417689.1
ENST00000561697.1 ENST00000317091.4 ENST00000566182.1 |
CES2
|
carboxylesterase 2 |
| chrX_+_146993449 | 0.21 |
ENST00000218200.8
ENST00000370471.3 ENST00000370477.1 |
FMR1
|
fragile X mental retardation 1 |
| chr19_+_18284477 | 0.20 |
ENST00000407280.3
|
IFI30
|
interferon, gamma-inducible protein 30 |
| chr22_+_31892373 | 0.20 |
ENST00000443011.1
ENST00000400289.1 ENST00000444859.1 ENST00000400288.2 |
SFI1
|
Sfi1 homolog, spindle assembly associated (yeast) |
| chr4_-_87855851 | 0.18 |
ENST00000473559.1
|
C4orf36
|
chromosome 4 open reading frame 36 |
| chr7_-_122526799 | 0.18 |
ENST00000334010.7
ENST00000313070.7 |
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr1_+_25599018 | 0.17 |
ENST00000417538.2
ENST00000357542.4 ENST00000568195.1 ENST00000342055.5 ENST00000423810.2 |
RHD
|
Rh blood group, D antigen |
| chrX_+_146993534 | 0.17 |
ENST00000334557.6
ENST00000439526.2 ENST00000370475.4 |
FMR1
|
fragile X mental retardation 1 |
| chr4_+_124320665 | 0.16 |
ENST00000394339.2
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr14_+_86401039 | 0.16 |
ENST00000557195.1
|
CTD-2341M24.1
|
CTD-2341M24.1 |
| chr4_+_100495864 | 0.16 |
ENST00000265517.5
ENST00000422897.2 |
MTTP
|
microsomal triglyceride transfer protein |
| chr6_+_26440700 | 0.16 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr16_+_29127282 | 0.16 |
ENST00000562902.1
|
RP11-426C22.5
|
RP11-426C22.5 |
| chr1_+_25598872 | 0.16 |
ENST00000328664.4
|
RHD
|
Rh blood group, D antigen |
| chr4_+_156588350 | 0.15 |
ENST00000296518.7
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr1_+_25598989 | 0.15 |
ENST00000454452.2
|
RHD
|
Rh blood group, D antigen |
| chr12_+_113376249 | 0.15 |
ENST00000551007.1
ENST00000548514.1 |
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr11_-_93583697 | 0.14 |
ENST00000409977.1
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr7_-_122526499 | 0.14 |
ENST00000412584.2
|
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr6_-_2876744 | 0.14 |
ENST00000420981.2
|
RP11-420G6.4
|
RP11-420G6.4 |
| chr2_+_127413481 | 0.12 |
ENST00000259254.4
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr12_-_11339543 | 0.12 |
ENST00000334266.1
|
TAS2R42
|
taste receptor, type 2, member 42 |
| chr12_+_133614062 | 0.12 |
ENST00000540031.1
ENST00000536123.1 |
ZNF84
|
zinc finger protein 84 |
| chr2_+_113672770 | 0.12 |
ENST00000311328.2
|
IL37
|
interleukin 37 |
| chr6_+_134758827 | 0.12 |
ENST00000431422.1
|
LINC01010
|
long intergenic non-protein coding RNA 1010 |
| chr1_+_151129135 | 0.12 |
ENST00000602841.1
|
SCNM1
|
sodium channel modifier 1 |
| chr16_-_67970990 | 0.11 |
ENST00000358514.4
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr2_+_219125714 | 0.11 |
ENST00000522678.1
ENST00000519574.1 ENST00000521462.1 |
GPBAR1
|
G protein-coupled bile acid receptor 1 |
| chr20_+_20033158 | 0.10 |
ENST00000340348.6
ENST00000377309.2 ENST00000389656.3 ENST00000377306.1 ENST00000245957.5 ENST00000377303.2 ENST00000475466.1 |
C20orf26
|
chromosome 20 open reading frame 26 |
| chr4_+_156587853 | 0.10 |
ENST00000506455.1
ENST00000511108.1 |
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chrX_+_154610428 | 0.10 |
ENST00000354514.4
|
H2AFB2
|
H2A histone family, member B2 |
| chr14_+_73525229 | 0.09 |
ENST00000527432.1
ENST00000531500.1 ENST00000525321.1 ENST00000526754.1 |
RBM25
|
RNA binding motif protein 25 |
| chr2_-_119605253 | 0.09 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr4_+_87856129 | 0.09 |
ENST00000395146.4
ENST00000507468.1 |
AFF1
|
AF4/FMR2 family, member 1 |
| chr12_-_322821 | 0.09 |
ENST00000359674.4
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr2_+_127413704 | 0.08 |
ENST00000409836.3
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr8_+_70476088 | 0.08 |
ENST00000525999.1
|
SULF1
|
sulfatase 1 |
| chr14_+_24605389 | 0.07 |
ENST00000382708.3
ENST00000561435.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr5_+_140749803 | 0.07 |
ENST00000576222.1
|
PCDHGB3
|
protocadherin gamma subfamily B, 3 |
| chr4_+_156587979 | 0.07 |
ENST00000511507.1
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr6_+_42018251 | 0.07 |
ENST00000372978.3
ENST00000494547.1 ENST00000456846.2 ENST00000372982.4 ENST00000472818.1 ENST00000372977.3 |
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr4_+_156588115 | 0.07 |
ENST00000455639.2
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr7_+_134832808 | 0.06 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr16_+_75681650 | 0.06 |
ENST00000300086.4
|
TERF2IP
|
telomeric repeat binding factor 2, interacting protein |
| chr4_-_74847800 | 0.06 |
ENST00000296029.3
|
PF4
|
platelet factor 4 |
| chr4_+_156588249 | 0.05 |
ENST00000393832.3
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr12_+_12223867 | 0.04 |
ENST00000308721.5
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr10_-_22292675 | 0.04 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr3_+_183815318 | 0.04 |
ENST00000425359.2
|
HTR3E
|
5-hydroxytryptamine (serotonin) receptor 3E, ionotropic |
| chr6_+_26402465 | 0.03 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr2_-_99485825 | 0.03 |
ENST00000423771.1
|
KIAA1211L
|
KIAA1211-like |
| chr2_-_175629135 | 0.02 |
ENST00000409542.1
ENST00000409219.1 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr6_-_711395 | 0.02 |
ENST00000606285.1
|
RP11-532F6.3
|
RP11-532F6.3 |
| chr2_-_175629164 | 0.02 |
ENST00000409323.1
ENST00000261007.5 ENST00000348749.5 |
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr7_-_126892303 | 0.01 |
ENST00000358373.3
|
GRM8
|
glutamate receptor, metabotropic 8 |
| chr12_+_12224331 | 0.01 |
ENST00000396367.1
ENST00000266434.4 ENST00000396369.1 |
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr20_-_56195525 | 0.00 |
ENST00000371173.3
ENST00000395822.3 ENST00000340462.4 ENST00000343535.4 |
ZBP1
|
Z-DNA binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 1.4 | 6.9 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 1.1 | 3.3 | GO:1901253 | negative regulation of dendritic cell cytokine production(GO:0002731) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.9 | 11.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.9 | 13.3 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.8 | 8.8 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.6 | 2.3 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.4 | 0.4 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.4 | 1.3 | GO:0002519 | natural killer cell tolerance induction(GO:0002519) positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.4 | 5.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.3 | 2.4 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.3 | 4.7 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.3 | 2.5 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.2 | 0.7 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.2 | 0.5 | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib(GO:0002428) |
| 0.2 | 0.9 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.2 | 0.9 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.2 | 0.6 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.2 | 4.1 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.2 | 1.5 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 | 1.7 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.2 | 1.6 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 1.6 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 1.0 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.8 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.7 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.1 | 0.5 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 3.4 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.1 | 6.0 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.1 | 0.1 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.1 | 1.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.8 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 3.8 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.1 | 2.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.6 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 2.4 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.1 | 0.4 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.1 | 0.6 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 2.1 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.1 | 1.4 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 1.1 | GO:0030202 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.4 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.1 | 0.9 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.1 | 5.8 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.1 | 0.6 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.6 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.5 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.2 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.0 | 1.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 2.8 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.4 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 1.6 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.9 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.2 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.8 | GO:0070206 | protein trimerization(GO:0070206) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.3 | 2.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.3 | 0.5 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.3 | 1.6 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 0.7 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.2 | 1.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 2.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 6.0 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.2 | 1.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 2.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.9 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.4 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.1 | 0.5 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 0.5 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 3.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 1.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.7 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 3.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.4 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 2.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 12.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 9.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.6 | 7.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.3 | 2.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.3 | 3.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.3 | 1.3 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.3 | 1.4 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.3 | 1.4 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.2 | 1.0 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) TAP binding(GO:0046977) TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.2 | 5.8 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.2 | 0.9 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.2 | 1.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 5.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 2.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 1.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.9 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.7 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 3.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.5 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.4 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 0.4 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.1 | 3.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.6 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 6.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 7.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 2.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 1.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 1.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 6.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 1.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 2.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 1.6 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.5 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 6.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 2.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.5 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 3.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 13.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.4 | 40.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 0.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 3.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 5.7 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 1.6 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 2.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.4 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 1.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |