Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
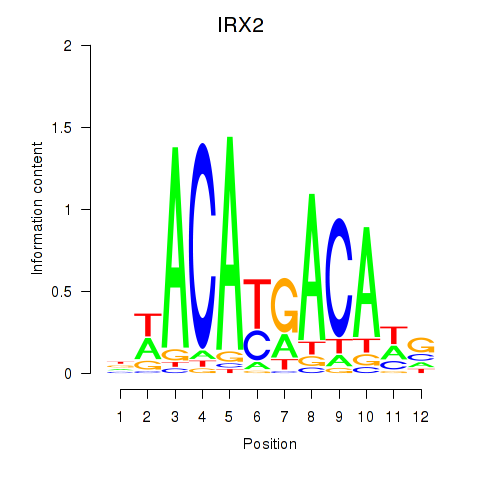
Results for IRX2
Z-value: 0.31
Transcription factors associated with IRX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX2
|
ENSG00000170561.8 | iroquois homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX2 | hg19_v2_chr5_-_2751762_2751784 | 0.18 | 4.4e-01 | Click! |
Activity profile of IRX2 motif
Sorted Z-values of IRX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_22033392 | 0.60 |
ENST00000320220.9
ENST00000415724.1 ENST00000422251.1 ENST00000382466.3 ENST00000542645.1 ENST00000400590.3 |
ZDHHC20
|
zinc finger, DHHC-type containing 20 |
| chr10_-_14596140 | 0.44 |
ENST00000496330.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr18_-_33702078 | 0.43 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr1_+_215747118 | 0.43 |
ENST00000448333.1
|
KCTD3
|
potassium channel tetramerization domain containing 3 |
| chr3_+_171844762 | 0.38 |
ENST00000443501.1
|
FNDC3B
|
fibronectin type III domain containing 3B |
| chr20_+_2083540 | 0.37 |
ENST00000400064.3
|
STK35
|
serine/threonine kinase 35 |
| chr4_+_169575875 | 0.35 |
ENST00000503457.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr8_-_90993869 | 0.35 |
ENST00000517772.1
|
NBN
|
nibrin |
| chr5_+_138210919 | 0.34 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr10_-_14574705 | 0.32 |
ENST00000489100.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr5_-_94417562 | 0.31 |
ENST00000505465.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr6_-_160209471 | 0.31 |
ENST00000539948.1
|
TCP1
|
t-complex 1 |
| chr8_-_29939933 | 0.30 |
ENST00000522794.1
|
TMEM66
|
transmembrane protein 66 |
| chr8_-_124428569 | 0.30 |
ENST00000521903.1
|
ATAD2
|
ATPase family, AAA domain containing 2 |
| chr5_-_142814241 | 0.30 |
ENST00000504572.1
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr4_-_22444733 | 0.28 |
ENST00000508133.1
|
GPR125
|
G protein-coupled receptor 125 |
| chr8_+_98900132 | 0.27 |
ENST00000520016.1
|
MATN2
|
matrilin 2 |
| chr7_-_21985489 | 0.24 |
ENST00000356195.5
ENST00000447180.1 ENST00000373934.4 ENST00000457951.1 |
CDCA7L
|
cell division cycle associated 7-like |
| chr5_+_158690089 | 0.24 |
ENST00000296786.6
|
UBLCP1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr7_+_16700806 | 0.24 |
ENST00000446596.1
ENST00000438834.1 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr10_-_14614311 | 0.23 |
ENST00000479731.1
ENST00000468492.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr12_+_112856690 | 0.23 |
ENST00000392597.1
ENST00000351677.2 |
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11 |
| chr16_+_58426296 | 0.23 |
ENST00000426538.2
ENST00000328514.7 ENST00000318129.5 |
GINS3
|
GINS complex subunit 3 (Psf3 homolog) |
| chr13_-_24007815 | 0.21 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chrX_+_9431324 | 0.21 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr11_-_33795893 | 0.21 |
ENST00000526785.1
ENST00000534136.1 ENST00000265651.3 ENST00000530401.1 ENST00000448981.2 |
FBXO3
|
F-box protein 3 |
| chr20_-_35329063 | 0.21 |
ENST00000422536.1
|
NDRG3
|
NDRG family member 3 |
| chr3_-_185641681 | 0.21 |
ENST00000259043.7
|
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chr2_-_3504587 | 0.20 |
ENST00000415131.1
|
ADI1
|
acireductone dioxygenase 1 |
| chr6_+_24775153 | 0.19 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr12_+_53773944 | 0.18 |
ENST00000551969.1
ENST00000327443.4 |
SP1
|
Sp1 transcription factor |
| chr6_-_136847610 | 0.18 |
ENST00000454590.1
ENST00000432797.2 |
MAP7
|
microtubule-associated protein 7 |
| chr5_-_94417339 | 0.18 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr11_+_62648357 | 0.18 |
ENST00000541372.1
ENST00000539458.1 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr10_+_46997926 | 0.18 |
ENST00000374314.4
|
GPRIN2
|
G protein regulated inducer of neurite outgrowth 2 |
| chr4_-_83821676 | 0.18 |
ENST00000355196.2
ENST00000507676.1 ENST00000506495.1 ENST00000507051.1 |
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr6_-_136847099 | 0.17 |
ENST00000438100.2
|
MAP7
|
microtubule-associated protein 7 |
| chr12_+_66218212 | 0.16 |
ENST00000393578.3
ENST00000425208.2 ENST00000536545.1 ENST00000354636.3 |
HMGA2
|
high mobility group AT-hook 2 |
| chr16_+_81272287 | 0.16 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr12_+_65996599 | 0.16 |
ENST00000539116.1
ENST00000541391.1 |
RP11-221N13.3
|
RP11-221N13.3 |
| chr16_-_71323617 | 0.16 |
ENST00000563876.1
|
CMTR2
|
cap methyltransferase 2 |
| chr2_+_27851863 | 0.16 |
ENST00000264718.3
ENST00000610189.1 |
GPN1
|
GPN-loop GTPase 1 |
| chr5_-_150138246 | 0.16 |
ENST00000518015.1
|
DCTN4
|
dynactin 4 (p62) |
| chrX_-_24045303 | 0.16 |
ENST00000328046.8
|
KLHL15
|
kelch-like family member 15 |
| chr4_-_71705060 | 0.16 |
ENST00000514161.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr1_+_206516200 | 0.16 |
ENST00000295713.5
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr4_+_48807155 | 0.15 |
ENST00000504654.1
|
OCIAD1
|
OCIA domain containing 1 |
| chr3_+_184534994 | 0.15 |
ENST00000441141.1
ENST00000445089.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr12_+_66218903 | 0.15 |
ENST00000393577.3
|
HMGA2
|
high mobility group AT-hook 2 |
| chr7_-_105332084 | 0.15 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr17_+_76037081 | 0.15 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr1_-_92371839 | 0.15 |
ENST00000370399.2
|
TGFBR3
|
transforming growth factor, beta receptor III |
| chr4_-_71705027 | 0.15 |
ENST00000545193.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr5_-_150138061 | 0.14 |
ENST00000521533.1
ENST00000424236.1 |
DCTN4
|
dynactin 4 (p62) |
| chr3_-_127441406 | 0.14 |
ENST00000487473.1
ENST00000484451.1 |
MGLL
|
monoglyceride lipase |
| chr15_-_45670924 | 0.14 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chrX_-_118739835 | 0.14 |
ENST00000542113.1
ENST00000304449.5 |
NKRF
|
NFKB repressing factor |
| chr10_+_90660832 | 0.14 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr6_+_37897735 | 0.14 |
ENST00000373389.5
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr4_+_26165074 | 0.14 |
ENST00000512351.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr15_+_57998923 | 0.13 |
ENST00000380557.4
|
POLR2M
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr17_-_5322786 | 0.13 |
ENST00000225696.4
|
NUP88
|
nucleoporin 88kDa |
| chr5_+_156696362 | 0.12 |
ENST00000377576.3
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr13_+_35516390 | 0.12 |
ENST00000540320.1
ENST00000400445.3 ENST00000310336.4 |
NBEA
|
neurobeachin |
| chr5_-_94417186 | 0.12 |
ENST00000312216.8
ENST00000512425.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr16_+_58537737 | 0.12 |
ENST00000561738.1
|
NDRG4
|
NDRG family member 4 |
| chr3_-_11645925 | 0.12 |
ENST00000413604.1
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr1_-_173793458 | 0.12 |
ENST00000356198.2
|
CENPL
|
centromere protein L |
| chr1_-_149783914 | 0.12 |
ENST00000369167.1
ENST00000427880.2 ENST00000545683.1 |
HIST2H2BF
|
histone cluster 2, H2bf |
| chr15_+_44580955 | 0.11 |
ENST00000345795.2
ENST00000360824.3 |
CASC4
|
cancer susceptibility candidate 4 |
| chr12_-_102513843 | 0.11 |
ENST00000551744.2
ENST00000552283.1 |
NUP37
|
nucleoporin 37kDa |
| chr8_+_40018977 | 0.11 |
ENST00000520487.1
|
RP11-470M17.2
|
RP11-470M17.2 |
| chr14_+_52327109 | 0.11 |
ENST00000335281.4
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr14_+_52327350 | 0.11 |
ENST00000555472.1
ENST00000556766.1 |
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr10_+_70320413 | 0.11 |
ENST00000373644.4
|
TET1
|
tet methylcytosine dioxygenase 1 |
| chr12_+_78359999 | 0.11 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr17_-_2615031 | 0.11 |
ENST00000576885.1
ENST00000574426.2 |
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr14_-_50999190 | 0.11 |
ENST00000557390.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr1_-_111506562 | 0.10 |
ENST00000485275.2
ENST00000369763.4 |
LRIF1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr3_-_15140629 | 0.10 |
ENST00000507357.1
ENST00000449050.1 ENST00000253699.3 ENST00000435849.3 ENST00000476527.2 |
ZFYVE20
|
zinc finger, FYVE domain containing 20 |
| chr10_+_102672712 | 0.10 |
ENST00000370271.3
ENST00000370269.3 ENST00000609386.1 |
FAM178A
|
family with sequence similarity 178, member A |
| chr4_+_114214125 | 0.10 |
ENST00000509550.1
|
ANK2
|
ankyrin 2, neuronal |
| chr6_-_27841289 | 0.10 |
ENST00000355981.2
|
HIST1H4L
|
histone cluster 1, H4l |
| chr4_+_57371509 | 0.09 |
ENST00000360096.2
|
ARL9
|
ADP-ribosylation factor-like 9 |
| chr1_+_10292308 | 0.09 |
ENST00000377081.1
|
KIF1B
|
kinesin family member 1B |
| chr4_-_38806404 | 0.09 |
ENST00000308979.2
ENST00000505940.1 ENST00000515861.1 |
TLR1
|
toll-like receptor 1 |
| chr22_+_29168652 | 0.09 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr5_+_167913450 | 0.09 |
ENST00000231572.3
ENST00000538719.1 |
RARS
|
arginyl-tRNA synthetase |
| chr7_+_72395617 | 0.09 |
ENST00000434423.2
|
POM121
|
POM121 transmembrane nucleoporin |
| chr9_-_13175823 | 0.09 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chr1_-_207095324 | 0.08 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr4_+_159131596 | 0.08 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr16_-_25026641 | 0.08 |
ENST00000289968.6
ENST00000303665.5 ENST00000455311.2 ENST00000441763.2 |
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr3_+_31574189 | 0.08 |
ENST00000295770.2
|
STT3B
|
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr11_+_62648336 | 0.08 |
ENST00000338663.7
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr4_-_14889791 | 0.08 |
ENST00000509654.1
ENST00000515031.1 ENST00000505089.2 |
LINC00504
|
long intergenic non-protein coding RNA 504 |
| chr14_-_65289812 | 0.08 |
ENST00000389720.3
ENST00000389721.5 ENST00000389722.3 |
SPTB
|
spectrin, beta, erythrocytic |
| chrX_-_148676974 | 0.08 |
ENST00000524178.1
|
HSFX2
|
heat shock transcription factor family, X linked 2 |
| chr17_+_7761013 | 0.08 |
ENST00000571846.1
|
CYB5D1
|
cytochrome b5 domain containing 1 |
| chr7_+_93652116 | 0.08 |
ENST00000415536.1
|
AC003092.1
|
AC003092.1 |
| chr5_-_94417314 | 0.08 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr14_+_58797974 | 0.08 |
ENST00000417477.2
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr12_+_54384370 | 0.07 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chr14_+_75536335 | 0.07 |
ENST00000554763.1
ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr19_+_13134772 | 0.07 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr1_+_152691998 | 0.07 |
ENST00000368775.2
|
C1orf68
|
chromosome 1 open reading frame 68 |
| chrX_+_148855726 | 0.07 |
ENST00000370416.4
|
HSFX1
|
heat shock transcription factor family, X linked 1 |
| chr2_+_113479063 | 0.07 |
ENST00000327581.4
|
NT5DC4
|
5'-nucleotidase domain containing 4 |
| chr17_+_44590076 | 0.06 |
ENST00000333412.3
|
LRRC37A2
|
leucine rich repeat containing 37, member A2 |
| chr11_+_33037652 | 0.06 |
ENST00000311388.3
|
DEPDC7
|
DEP domain containing 7 |
| chr4_+_119606523 | 0.06 |
ENST00000388822.5
ENST00000506780.1 ENST00000508801.1 |
METTL14
|
methyltransferase like 14 |
| chr17_+_7761301 | 0.06 |
ENST00000332439.4
ENST00000570446.1 |
CYB5D1
|
cytochrome b5 domain containing 1 |
| chr4_+_159131630 | 0.06 |
ENST00000504569.1
ENST00000509278.1 ENST00000514558.1 ENST00000503200.1 |
TMEM144
|
transmembrane protein 144 |
| chr3_-_194072019 | 0.06 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr4_-_153332886 | 0.06 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr1_-_151148442 | 0.06 |
ENST00000441701.1
ENST00000416280.2 |
TMOD4
|
tropomodulin 4 (muscle) |
| chr1_+_110527308 | 0.06 |
ENST00000369799.5
|
AHCYL1
|
adenosylhomocysteinase-like 1 |
| chr14_+_50999744 | 0.05 |
ENST00000441560.2
|
ATL1
|
atlastin GTPase 1 |
| chr16_+_82068585 | 0.05 |
ENST00000563491.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr9_+_35792151 | 0.05 |
ENST00000342694.2
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
| chr7_-_33140498 | 0.05 |
ENST00000448915.1
|
RP9
|
retinitis pigmentosa 9 (autosomal dominant) |
| chr4_+_88896819 | 0.05 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr8_+_67405794 | 0.04 |
ENST00000522977.1
ENST00000480005.1 |
C8orf46
|
chromosome 8 open reading frame 46 |
| chr6_+_30908747 | 0.04 |
ENST00000462446.1
ENST00000304311.2 |
DPCR1
|
diffuse panbronchiolitis critical region 1 |
| chr2_+_159651821 | 0.04 |
ENST00000309950.3
ENST00000409042.1 |
DAPL1
|
death associated protein-like 1 |
| chr18_-_53070913 | 0.04 |
ENST00000568186.1
ENST00000564228.1 |
TCF4
|
transcription factor 4 |
| chr2_+_46844290 | 0.04 |
ENST00000238892.3
|
CRIPT
|
cysteine-rich PDZ-binding protein |
| chrX_-_119077695 | 0.03 |
ENST00000371410.3
|
NKAP
|
NFKB activating protein |
| chr12_+_40787194 | 0.03 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr19_-_29704448 | 0.03 |
ENST00000304863.4
|
UQCRFS1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr17_+_34842473 | 0.03 |
ENST00000490126.2
ENST00000225410.4 |
ZNHIT3
|
zinc finger, HIT-type containing 3 |
| chr16_+_57680043 | 0.03 |
ENST00000569154.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr8_-_124665190 | 0.02 |
ENST00000325995.7
|
KLHL38
|
kelch-like family member 38 |
| chr6_+_27925019 | 0.02 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr8_+_81397846 | 0.02 |
ENST00000379091.4
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr9_+_79792410 | 0.02 |
ENST00000357409.5
|
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae) |
| chr1_-_54405773 | 0.02 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr12_+_32655110 | 0.02 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr16_+_69373323 | 0.02 |
ENST00000254940.5
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr3_-_81792780 | 0.02 |
ENST00000489715.1
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr2_-_233877912 | 0.02 |
ENST00000264051.3
|
NGEF
|
neuronal guanine nucleotide exchange factor |
| chr2_-_46844242 | 0.02 |
ENST00000281382.6
|
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr17_+_38121772 | 0.02 |
ENST00000577447.1
|
GSDMA
|
gasdermin A |
| chr3_+_169539710 | 0.02 |
ENST00000340806.6
|
LRRIQ4
|
leucine-rich repeats and IQ motif containing 4 |
| chr1_-_207095212 | 0.02 |
ENST00000420007.2
|
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chrX_-_70331298 | 0.02 |
ENST00000456850.2
ENST00000473378.1 ENST00000487883.1 ENST00000374202.2 |
IL2RG
|
interleukin 2 receptor, gamma |
| chr8_-_11996586 | 0.01 |
ENST00000333796.3
|
USP17L2
|
ubiquitin specific peptidase 17-like family member 2 |
| chr14_+_75536280 | 0.01 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr19_-_55953704 | 0.01 |
ENST00000416792.1
|
SHISA7
|
shisa family member 7 |
| chr2_-_109605663 | 0.01 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr1_+_204839959 | 0.01 |
ENST00000404076.1
|
NFASC
|
neurofascin |
| chrX_-_117119243 | 0.01 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr1_-_151148492 | 0.01 |
ENST00000295314.4
|
TMOD4
|
tropomodulin 4 (muscle) |
| chr17_-_41322332 | 0.01 |
ENST00000590740.1
|
RP11-242D8.1
|
RP11-242D8.1 |
| chr17_-_60885645 | 0.01 |
ENST00000544856.2
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr11_-_110561721 | 0.01 |
ENST00000357139.3
|
ARHGAP20
|
Rho GTPase activating protein 20 |
| chr3_+_148457585 | 0.01 |
ENST00000402260.1
|
AGTR1
|
angiotensin II receptor, type 1 |
| chr5_-_180242576 | 0.01 |
ENST00000514438.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr9_+_130159593 | 0.01 |
ENST00000419132.1
|
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr3_-_122102065 | 0.01 |
ENST00000479899.1
ENST00000291458.5 ENST00000497726.1 |
CCDC58
|
coiled-coil domain containing 58 |
| chrX_-_138790348 | 0.01 |
ENST00000414978.1
ENST00000519895.1 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr1_+_206317450 | 0.01 |
ENST00000358184.2
ENST00000361052.3 ENST00000360218.2 |
CTSE
|
cathepsin E |
| chr15_+_81391740 | 0.01 |
ENST00000561216.1
|
C15orf26
|
chromosome 15 open reading frame 26 |
| chr17_-_27054952 | 0.00 |
ENST00000580518.1
|
TLCD1
|
TLC domain containing 1 |
| chrX_+_155227246 | 0.00 |
ENST00000244174.5
ENST00000424344.3 |
IL9R
|
interleukin 9 receptor |
| chr4_-_168155417 | 0.00 |
ENST00000511269.1
ENST00000506697.1 ENST00000512042.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr15_+_38226827 | 0.00 |
ENST00000559502.1
ENST00000558148.1 ENST00000558158.1 |
TMCO5A
|
transmembrane and coiled-coil domains 5A |
| chr19_-_51334769 | 0.00 |
ENST00000596931.1
ENST00000416184.1 ENST00000301421.2 ENST00000598239.1 |
KLK15
|
kallikrein-related peptidase 15 |
| chr10_+_1095416 | 0.00 |
ENST00000358220.1
|
WDR37
|
WD repeat domain 37 |
| chr2_-_8715616 | 0.00 |
ENST00000418358.1
|
AC011747.3
|
AC011747.3 |
| chr1_+_206317591 | 0.00 |
ENST00000432969.2
|
CTSE
|
cathepsin E |
| chr17_+_74075263 | 0.00 |
ENST00000334586.5
ENST00000392503.2 |
ZACN
|
zinc activated ligand-gated ion channel |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0044417 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.1 | 0.3 | GO:0003131 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.1 | 0.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.2 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.1 | 0.3 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.2 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.0 | 0.2 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.2 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.3 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.1 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.1 | GO:0034130 | toll-like receptor 1 signaling pathway(GO:0034130) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.2 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 0.2 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.2 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.4 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.0 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.1 | 0.2 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.1 | 0.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.0 | 0.1 | GO:0015067 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.0 | 0.3 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |