Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
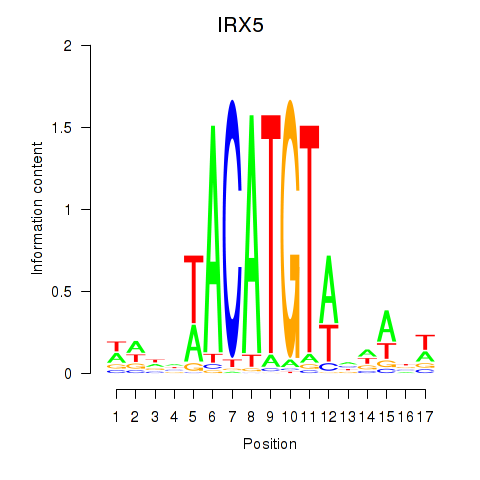
Results for IRX5
Z-value: 0.47
Transcription factors associated with IRX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX5
|
ENSG00000176842.10 | iroquois homeobox 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX5 | hg19_v2_chr16_+_54964740_54964789 | 0.35 | 1.3e-01 | Click! |
Activity profile of IRX5 motif
Sorted Z-values of IRX5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_141264597 | 1.96 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr13_+_97928395 | 0.97 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr10_-_115904361 | 0.72 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr4_-_104119528 | 0.70 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr5_+_169011033 | 0.69 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr8_+_27631903 | 0.67 |
ENST00000305188.8
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr15_+_66585555 | 0.60 |
ENST00000319194.5
ENST00000525134.2 ENST00000441424.2 |
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr8_+_22225041 | 0.55 |
ENST00000289952.5
ENST00000524285.1 |
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr15_-_64665911 | 0.55 |
ENST00000606793.1
ENST00000561349.1 ENST00000560278.1 |
CTD-2116N17.1
|
Uncharacterized protein |
| chr1_-_197115818 | 0.54 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr3_+_180586536 | 0.51 |
ENST00000465551.1
|
FXR1
|
fragile X mental retardation, autosomal homolog 1 |
| chr12_-_100486668 | 0.50 |
ENST00000550544.1
ENST00000551980.1 ENST00000548045.1 ENST00000545232.2 ENST00000551973.1 |
UHRF1BP1L
|
UHRF1 binding protein 1-like |
| chr1_+_212475148 | 0.50 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr7_-_55583740 | 0.49 |
ENST00000453256.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr14_+_73563735 | 0.48 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25 |
| chr4_+_169842707 | 0.48 |
ENST00000503290.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr1_+_220267429 | 0.46 |
ENST00000366922.1
ENST00000302637.5 |
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr3_-_27498235 | 0.45 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr3_-_124606502 | 0.44 |
ENST00000483168.1
|
ITGB5
|
integrin, beta 5 |
| chr5_+_169010638 | 0.44 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr2_+_191792376 | 0.42 |
ENST00000409428.1
ENST00000409215.1 |
GLS
|
glutaminase |
| chr21_+_38792602 | 0.41 |
ENST00000398960.2
ENST00000398956.2 |
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr8_-_101962777 | 0.40 |
ENST00000395951.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr18_+_22006646 | 0.39 |
ENST00000585067.1
ENST00000578221.1 |
IMPACT
|
impact RWD domain protein |
| chr11_+_61891445 | 0.39 |
ENST00000394818.3
ENST00000533896.1 ENST00000278849.4 |
INCENP
|
inner centromere protein antigens 135/155kDa |
| chr9_-_19065082 | 0.39 |
ENST00000415524.1
|
HAUS6
|
HAUS augmin-like complex, subunit 6 |
| chr4_-_104119488 | 0.38 |
ENST00000514974.1
|
CENPE
|
centromere protein E, 312kDa |
| chr2_+_160590469 | 0.38 |
ENST00000409591.1
|
MARCH7
|
membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase |
| chr2_+_113299990 | 0.37 |
ENST00000537335.1
ENST00000417433.2 |
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa |
| chr1_-_8763278 | 0.35 |
ENST00000468247.1
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr5_+_179135246 | 0.35 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr13_+_76362974 | 0.34 |
ENST00000497947.2
|
LMO7
|
LIM domain 7 |
| chr3_-_52713729 | 0.33 |
ENST00000296302.7
ENST00000356770.4 ENST00000337303.4 ENST00000409057.1 ENST00000410007.1 ENST00000409114.3 ENST00000409767.1 ENST00000423351.1 |
PBRM1
|
polybromo 1 |
| chr9_-_125667618 | 0.33 |
ENST00000423239.2
|
RC3H2
|
ring finger and CCCH-type domains 2 |
| chrX_-_15353629 | 0.33 |
ENST00000333590.4
ENST00000428964.1 ENST00000542278.1 |
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A |
| chr12_-_120315074 | 0.32 |
ENST00000261833.7
ENST00000392521.2 |
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr7_+_116502527 | 0.32 |
ENST00000361183.3
|
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr1_-_244006528 | 0.31 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr4_-_143481822 | 0.30 |
ENST00000510812.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr7_+_133615169 | 0.29 |
ENST00000541309.1
|
EXOC4
|
exocyst complex component 4 |
| chr17_-_47723943 | 0.29 |
ENST00000510476.1
ENST00000503676.1 |
SPOP
|
speckle-type POZ protein |
| chr4_-_22444733 | 0.29 |
ENST00000508133.1
|
GPR125
|
G protein-coupled receptor 125 |
| chr1_-_85156417 | 0.29 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr1_-_115124257 | 0.29 |
ENST00000369541.3
|
BCAS2
|
breast carcinoma amplified sequence 2 |
| chr17_-_6524159 | 0.28 |
ENST00000589033.1
|
KIAA0753
|
KIAA0753 |
| chr16_-_58585513 | 0.28 |
ENST00000245138.4
ENST00000567285.1 |
CNOT1
|
CCR4-NOT transcription complex, subunit 1 |
| chr3_+_111630451 | 0.28 |
ENST00000495180.1
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr12_+_58335360 | 0.27 |
ENST00000300145.3
|
XRCC6BP1
|
XRCC6 binding protein 1 |
| chr17_+_75447326 | 0.27 |
ENST00000591088.1
|
SEPT9
|
septin 9 |
| chr5_-_156390230 | 0.26 |
ENST00000407087.3
ENST00000274532.2 |
TIMD4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr13_+_24844857 | 0.26 |
ENST00000409126.1
ENST00000343003.6 |
SPATA13
|
spermatogenesis associated 13 |
| chr16_+_19535133 | 0.26 |
ENST00000396212.2
ENST00000381396.5 |
CCP110
|
centriolar coiled coil protein 110kDa |
| chr1_+_87458692 | 0.26 |
ENST00000370548.2
ENST00000356813.4 |
RP5-1052I5.2
HS2ST1
|
Heparan sulfate 2-O-sulfotransferase 1 heparan sulfate 2-O-sulfotransferase 1 |
| chrX_-_135962923 | 0.26 |
ENST00000565438.1
|
RBMX
|
RNA binding motif protein, X-linked |
| chr20_+_43990576 | 0.25 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr6_+_126221034 | 0.25 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chrX_+_12809463 | 0.25 |
ENST00000380663.3
ENST00000380668.5 ENST00000398491.2 ENST00000489404.1 |
PRPS2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr10_-_35379524 | 0.25 |
ENST00000374751.3
ENST00000374742.1 ENST00000602371.1 |
CUL2
|
cullin 2 |
| chr2_+_38177575 | 0.25 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr4_+_75480629 | 0.25 |
ENST00000380846.3
|
AREGB
|
amphiregulin B |
| chr4_+_41361616 | 0.25 |
ENST00000513024.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr14_+_56127960 | 0.25 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr12_-_123728548 | 0.24 |
ENST00000545406.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr10_+_90660832 | 0.24 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr14_-_68159152 | 0.24 |
ENST00000554035.1
|
RDH11
|
retinol dehydrogenase 11 (all-trans/9-cis/11-cis) |
| chr1_-_227505289 | 0.23 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr7_+_77469439 | 0.23 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr3_+_158787041 | 0.23 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr5_-_93447333 | 0.22 |
ENST00000395965.3
ENST00000505869.1 ENST00000509163.1 |
FAM172A
|
family with sequence similarity 172, member A |
| chr16_+_19535235 | 0.22 |
ENST00000565376.2
ENST00000396208.2 |
CCP110
|
centriolar coiled coil protein 110kDa |
| chr1_+_63989004 | 0.22 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr5_+_54455946 | 0.22 |
ENST00000503787.1
ENST00000296734.6 ENST00000515370.1 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr8_-_108510224 | 0.21 |
ENST00000517746.1
ENST00000297450.3 |
ANGPT1
|
angiopoietin 1 |
| chr3_-_127541194 | 0.21 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chr14_+_56127989 | 0.21 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr10_-_91403625 | 0.21 |
ENST00000322191.6
ENST00000342512.3 ENST00000371774.2 |
PANK1
|
pantothenate kinase 1 |
| chr11_+_57531292 | 0.21 |
ENST00000524579.1
|
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr15_+_66585879 | 0.21 |
ENST00000319212.4
|
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr5_+_140566 | 0.20 |
ENST00000502646.1
|
PLEKHG4B
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4B |
| chr11_+_34999328 | 0.20 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr15_+_81225699 | 0.20 |
ENST00000560027.1
|
KIAA1199
|
KIAA1199 |
| chr4_+_75310851 | 0.20 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr12_-_25102252 | 0.20 |
ENST00000261192.7
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chrX_+_108779004 | 0.20 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr14_+_51706886 | 0.20 |
ENST00000457354.2
|
TMX1
|
thioredoxin-related transmembrane protein 1 |
| chr6_-_111804905 | 0.20 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr10_+_86184676 | 0.19 |
ENST00000543283.1
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr6_-_111804393 | 0.19 |
ENST00000368802.3
ENST00000368805.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr3_-_196987309 | 0.18 |
ENST00000453607.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr15_+_49170083 | 0.18 |
ENST00000530028.2
|
EID1
|
EP300 interacting inhibitor of differentiation 1 |
| chr1_-_149859466 | 0.18 |
ENST00000331128.3
|
HIST2H2AB
|
histone cluster 2, H2ab |
| chr4_+_159131630 | 0.18 |
ENST00000504569.1
ENST00000509278.1 ENST00000514558.1 ENST00000503200.1 |
TMEM144
|
transmembrane protein 144 |
| chr18_+_22006580 | 0.17 |
ENST00000284202.4
|
IMPACT
|
impact RWD domain protein |
| chr13_+_24844819 | 0.17 |
ENST00000399949.2
|
SPATA13
|
spermatogenesis associated 13 |
| chr1_-_23504176 | 0.17 |
ENST00000302291.4
|
LUZP1
|
leucine zipper protein 1 |
| chr3_-_149470229 | 0.17 |
ENST00000473414.1
|
COMMD2
|
COMM domain containing 2 |
| chr19_-_36909528 | 0.17 |
ENST00000392161.3
ENST00000392171.1 |
ZFP82
|
ZFP82 zinc finger protein |
| chr19_-_44952635 | 0.17 |
ENST00000592308.1
ENST00000588931.1 ENST00000291187.4 |
ZNF229
|
zinc finger protein 229 |
| chr1_+_97188188 | 0.16 |
ENST00000541987.1
|
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr19_+_50691437 | 0.16 |
ENST00000598205.1
|
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr1_-_85870177 | 0.16 |
ENST00000542148.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr13_-_41768654 | 0.16 |
ENST00000379483.3
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7 |
| chr12_-_76462713 | 0.16 |
ENST00000552056.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr4_+_26344754 | 0.15 |
ENST00000515573.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr2_+_201173667 | 0.15 |
ENST00000409755.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr13_+_50589390 | 0.15 |
ENST00000360473.4
ENST00000312942.1 |
KCNRG
|
potassium channel regulator |
| chr4_-_75695366 | 0.14 |
ENST00000512743.1
|
BTC
|
betacellulin |
| chr17_+_38673270 | 0.14 |
ENST00000578280.1
|
RP5-1028K7.2
|
RP5-1028K7.2 |
| chr1_+_240408560 | 0.14 |
ENST00000441342.1
ENST00000545751.1 |
FMN2
|
formin 2 |
| chr22_+_20877924 | 0.14 |
ENST00000445189.1
|
MED15
|
mediator complex subunit 15 |
| chr11_+_110225855 | 0.14 |
ENST00000526605.1
ENST00000526703.1 |
RP11-347E10.1
|
RP11-347E10.1 |
| chr16_+_19467772 | 0.14 |
ENST00000219821.5
ENST00000561503.1 ENST00000564959.1 |
TMC5
|
transmembrane channel-like 5 |
| chr12_-_102224704 | 0.14 |
ENST00000299314.7
|
GNPTAB
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr4_+_144312659 | 0.14 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr17_+_41561317 | 0.14 |
ENST00000540306.1
ENST00000262415.3 ENST00000605777.1 |
DHX8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr2_-_32236002 | 0.13 |
ENST00000404530.1
|
MEMO1
|
mediator of cell motility 1 |
| chr8_+_27947746 | 0.13 |
ENST00000521015.1
ENST00000521570.1 |
ELP3
|
elongator acetyltransferase complex subunit 3 |
| chr7_-_111424506 | 0.13 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr22_+_18721427 | 0.13 |
ENST00000342888.3
|
AC008132.1
|
Uncharacterized protein |
| chr6_+_12008986 | 0.12 |
ENST00000491710.1
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr14_+_72052983 | 0.12 |
ENST00000358550.2
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr4_+_159131596 | 0.12 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr18_+_33709834 | 0.11 |
ENST00000358232.6
ENST00000351393.6 ENST00000442325.2 ENST00000423854.2 ENST00000350494.6 ENST00000542824.1 |
ELP2
|
elongator acetyltransferase complex subunit 2 |
| chr7_-_87856280 | 0.11 |
ENST00000490437.1
ENST00000431660.1 |
SRI
|
sorcin |
| chr2_-_242041607 | 0.11 |
ENST00000434791.1
ENST00000401626.2 ENST00000439144.1 ENST00000406593.1 ENST00000495694.1 ENST00000407095.3 ENST00000391980.2 |
MTERFD2
|
MTERF domain containing 2 |
| chr9_+_96846740 | 0.11 |
ENST00000288976.3
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chr5_+_67576109 | 0.11 |
ENST00000522084.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr15_-_56757329 | 0.11 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr14_-_95624227 | 0.11 |
ENST00000526495.1
|
DICER1
|
dicer 1, ribonuclease type III |
| chr19_-_52531600 | 0.11 |
ENST00000356322.6
ENST00000270649.6 |
ZNF614
|
zinc finger protein 614 |
| chr10_-_15130767 | 0.11 |
ENST00000356189.5
|
ACBD7
|
acyl-CoA binding domain containing 7 |
| chr17_+_12859080 | 0.10 |
ENST00000583608.1
|
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr12_+_106751436 | 0.10 |
ENST00000228347.4
|
POLR3B
|
polymerase (RNA) III (DNA directed) polypeptide B |
| chr6_+_26204825 | 0.10 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr7_-_87856303 | 0.10 |
ENST00000394641.3
|
SRI
|
sorcin |
| chr3_+_186158169 | 0.10 |
ENST00000435548.1
ENST00000421006.1 |
RP11-78H24.1
|
RP11-78H24.1 |
| chr4_-_103998439 | 0.10 |
ENST00000503230.1
ENST00000503818.1 |
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr8_-_67976509 | 0.10 |
ENST00000518747.1
|
COPS5
|
COP9 signalosome subunit 5 |
| chr7_-_112430427 | 0.10 |
ENST00000449743.1
ENST00000441474.1 ENST00000454074.1 ENST00000447395.1 |
TMEM168
|
transmembrane protein 168 |
| chr10_-_14050522 | 0.10 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr5_+_136070614 | 0.10 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr4_+_113739244 | 0.09 |
ENST00000503271.1
ENST00000503423.1 ENST00000506722.1 |
ANK2
|
ankyrin 2, neuronal |
| chr8_+_125463048 | 0.09 |
ENST00000328599.3
|
TRMT12
|
tRNA methyltransferase 12 homolog (S. cerevisiae) |
| chr3_-_72897545 | 0.09 |
ENST00000325599.8
|
SHQ1
|
SHQ1, H/ACA ribonucleoprotein assembly factor |
| chr1_-_145826450 | 0.09 |
ENST00000462900.2
|
GPR89A
|
G protein-coupled receptor 89A |
| chr18_-_69246167 | 0.09 |
ENST00000566582.1
|
RP11-510D19.1
|
RP11-510D19.1 |
| chr7_+_871559 | 0.08 |
ENST00000421580.1
|
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr8_+_101349823 | 0.08 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr4_+_189376725 | 0.08 |
ENST00000510832.1
ENST00000510005.1 ENST00000503458.1 |
LINC01060
|
long intergenic non-protein coding RNA 1060 |
| chr19_-_50266580 | 0.08 |
ENST00000246801.3
|
TSKS
|
testis-specific serine kinase substrate |
| chr8_+_67976593 | 0.08 |
ENST00000262210.5
ENST00000412460.1 |
CSPP1
|
centrosome and spindle pole associated protein 1 |
| chr15_+_21145765 | 0.08 |
ENST00000553416.1
|
CT60
|
cancer/testis antigen 60 (non-protein coding) |
| chr6_+_106988986 | 0.07 |
ENST00000457437.1
ENST00000535438.1 |
AIM1
|
absent in melanoma 1 |
| chr2_-_127963343 | 0.07 |
ENST00000335247.7
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr1_-_241683001 | 0.07 |
ENST00000366560.3
|
FH
|
fumarate hydratase |
| chr18_-_69246186 | 0.07 |
ENST00000568095.1
|
RP11-510D19.1
|
RP11-510D19.1 |
| chr7_-_111424462 | 0.07 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr11_+_31531291 | 0.07 |
ENST00000350638.5
ENST00000379163.5 ENST00000395934.2 |
ELP4
|
elongator acetyltransferase complex subunit 4 |
| chr1_+_53308398 | 0.07 |
ENST00000371528.1
|
ZYG11A
|
zyg-11 family member A, cell cycle regulator |
| chr12_+_32655110 | 0.07 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr2_-_40657397 | 0.06 |
ENST00000408028.2
ENST00000332839.4 ENST00000406391.2 ENST00000542024.1 ENST00000542756.1 ENST00000405901.3 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr2_+_173686303 | 0.06 |
ENST00000397087.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr20_+_54933971 | 0.06 |
ENST00000371384.3
ENST00000437418.1 |
FAM210B
|
family with sequence similarity 210, member B |
| chr20_-_29896388 | 0.06 |
ENST00000400549.1
|
DEFB116
|
defensin, beta 116 |
| chr18_-_44702668 | 0.06 |
ENST00000256433.3
|
IER3IP1
|
immediate early response 3 interacting protein 1 |
| chr1_-_21377447 | 0.06 |
ENST00000374937.3
ENST00000264211.8 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr9_-_3489428 | 0.06 |
ENST00000451859.1
|
RFX3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr15_-_44116873 | 0.06 |
ENST00000267812.3
|
MFAP1
|
microfibrillar-associated protein 1 |
| chr3_-_81792780 | 0.05 |
ENST00000489715.1
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr17_-_7108436 | 0.05 |
ENST00000493294.1
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr4_+_123653882 | 0.05 |
ENST00000433287.1
|
BBS12
|
Bardet-Biedl syndrome 12 |
| chr6_-_138833630 | 0.05 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr3_-_3221358 | 0.05 |
ENST00000424814.1
ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN
|
cereblon |
| chr11_+_114310237 | 0.05 |
ENST00000539119.1
|
REXO2
|
RNA exonuclease 2 |
| chrX_+_47867194 | 0.05 |
ENST00000376940.3
|
SPACA5
|
sperm acrosome associated 5 |
| chr10_-_51958906 | 0.05 |
ENST00000489640.1
|
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chrX_+_47990039 | 0.05 |
ENST00000304270.5
|
SPACA5B
|
sperm acrosome associated 5B |
| chr3_+_186383741 | 0.05 |
ENST00000232003.4
|
HRG
|
histidine-rich glycoprotein |
| chr14_+_35747825 | 0.05 |
ENST00000540871.1
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr4_+_122722466 | 0.05 |
ENST00000243498.5
ENST00000379663.3 ENST00000509800.1 |
EXOSC9
|
exosome component 9 |
| chr11_-_22647350 | 0.05 |
ENST00000327470.3
|
FANCF
|
Fanconi anemia, complementation group F |
| chr10_-_96829246 | 0.04 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr12_+_32655048 | 0.04 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr1_+_196857144 | 0.04 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr1_+_158969752 | 0.04 |
ENST00000566111.1
|
IFI16
|
interferon, gamma-inducible protein 16 |
| chr1_+_202317815 | 0.04 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chrX_-_83757399 | 0.04 |
ENST00000373177.2
ENST00000297977.5 ENST00000506585.2 ENST00000449553.2 |
HDX
|
highly divergent homeobox |
| chr4_+_113558612 | 0.04 |
ENST00000505034.1
ENST00000324052.6 |
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr1_+_53480598 | 0.04 |
ENST00000430330.2
ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2
|
sterol carrier protein 2 |
| chr18_-_32924372 | 0.04 |
ENST00000261332.6
ENST00000399061.3 |
ZNF24
|
zinc finger protein 24 |
| chr11_-_6790286 | 0.04 |
ENST00000338569.2
|
OR2AG2
|
olfactory receptor, family 2, subfamily AG, member 2 |
| chr16_+_56969284 | 0.04 |
ENST00000568358.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr14_+_39944025 | 0.03 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr10_-_18944123 | 0.03 |
ENST00000606425.1
|
RP11-139J15.7
|
Uncharacterized protein |
| chr9_-_37465396 | 0.03 |
ENST00000307750.4
|
ZBTB5
|
zinc finger and BTB domain containing 5 |
| chr17_+_14277419 | 0.03 |
ENST00000436469.1
|
AC022816.2
|
AC022816.2 |
| chrX_-_138790348 | 0.03 |
ENST00000414978.1
ENST00000519895.1 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr5_+_96079240 | 0.03 |
ENST00000515663.1
|
CAST
|
calpastatin |
| chr3_+_12392971 | 0.03 |
ENST00000287820.6
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr17_-_18430160 | 0.03 |
ENST00000392176.3
|
FAM106A
|
family with sequence similarity 106, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0071264 | regulation of eIF2 alpha phosphorylation by amino acid starvation(GO:0060733) regulation of translational initiation in response to starvation(GO:0071262) positive regulation of translational initiation in response to starvation(GO:0071264) |
| 0.2 | 0.5 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.2 | 1.1 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 0.5 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 0.5 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 2.0 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.7 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.5 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 1.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.2 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.3 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 0.2 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.1 | 0.3 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.3 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.3 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.8 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.2 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.4 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 2.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.6 | GO:1903874 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 0.2 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) Tie signaling pathway(GO:0048014) |
| 0.0 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.4 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.3 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.2 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.2 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.2 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:0055118 | negative regulation of heart rate(GO:0010459) negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.4 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.0 | 0.5 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.4 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.5 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.4 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.1 | 0.4 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.3 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.8 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.4 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.1 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.5 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.3 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.1 | 0.2 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.1 | 0.4 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.6 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.5 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.2 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.3 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.2 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 1.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) all-trans retinal binding(GO:0005503) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.4 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.0 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.3 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.1 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |