Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for IRX6_IRX4
Z-value: 0.50
Transcription factors associated with IRX6_IRX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
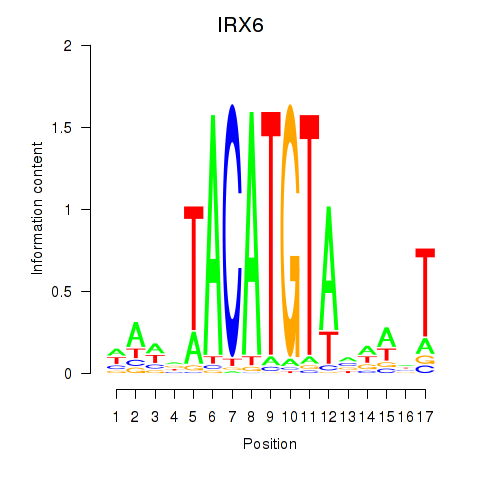
IRX6
|
ENSG00000159387.7 | iroquois homeobox 6 |
|
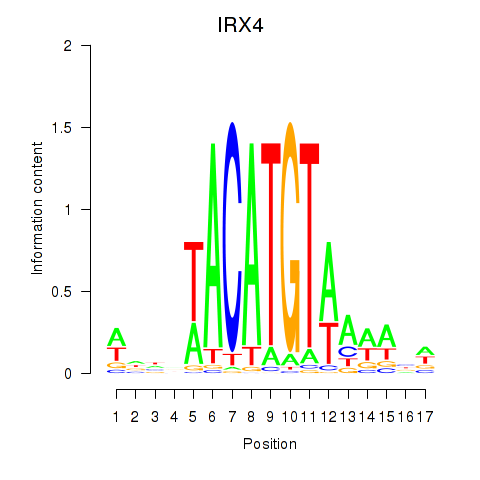
IRX4
|
ENSG00000113430.5 | iroquois homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX4 | hg19_v2_chr5_-_1882858_1883003 | -0.72 | 3.3e-04 | Click! |
| IRX6 | hg19_v2_chr16_+_55357672_55357672 | 0.01 | 9.6e-01 | Click! |
Activity profile of IRX6_IRX4 motif
Sorted Z-values of IRX6_IRX4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_16844611 | 1.62 |
ENST00000401412.1
ENST00000419304.2 |
AGR2
|
anterior gradient 2 |
| chr11_+_114166536 | 1.37 |
ENST00000299964.3
|
NNMT
|
nicotinamide N-methyltransferase |
| chr15_-_63448973 | 1.27 |
ENST00000462430.1
|
RPS27L
|
ribosomal protein S27-like |
| chr4_+_141264597 | 1.15 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr20_+_10199468 | 1.07 |
ENST00000254976.2
ENST00000304886.2 |
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr3_-_124606502 | 0.98 |
ENST00000483168.1
|
ITGB5
|
integrin, beta 5 |
| chr10_+_90660832 | 0.83 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr3_+_53528659 | 0.82 |
ENST00000350061.5
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr4_+_41361616 | 0.78 |
ENST00000513024.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_-_201438282 | 0.77 |
ENST00000367311.3
ENST00000367309.1 |
PHLDA3
|
pleckstrin homology-like domain, family A, member 3 |
| chr8_-_17555164 | 0.70 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr16_+_776936 | 0.69 |
ENST00000549114.1
ENST00000341413.4 ENST00000562187.1 ENST00000564537.1 |
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr16_+_777246 | 0.67 |
ENST00000561546.1
ENST00000564545.1 ENST00000389703.3 ENST00000567414.1 ENST00000568141.1 |
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr15_+_66585879 | 0.66 |
ENST00000319212.4
|
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr15_+_66585555 | 0.63 |
ENST00000319194.5
ENST00000525134.2 ENST00000441424.2 |
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr17_+_38673270 | 0.63 |
ENST00000578280.1
|
RP5-1028K7.2
|
RP5-1028K7.2 |
| chrX_-_135962923 | 0.60 |
ENST00000565438.1
|
RBMX
|
RNA binding motif protein, X-linked |
| chr3_-_9994021 | 0.58 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr12_-_76462713 | 0.52 |
ENST00000552056.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr8_+_27631903 | 0.49 |
ENST00000305188.8
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr1_-_89641680 | 0.48 |
ENST00000294671.2
|
GBP7
|
guanylate binding protein 7 |
| chr19_-_54974894 | 0.48 |
ENST00000333834.4
|
LENG9
|
leukocyte receptor cluster (LRC) member 9 |
| chr20_+_10199566 | 0.44 |
ENST00000430336.1
|
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr16_+_777739 | 0.38 |
ENST00000563792.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr2_-_231989808 | 0.37 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr12_-_123728548 | 0.35 |
ENST00000545406.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr6_-_33679452 | 0.34 |
ENST00000374231.4
ENST00000607484.1 ENST00000374214.3 |
UQCC2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr1_-_3566590 | 0.33 |
ENST00000424367.1
ENST00000378322.3 |
WRAP73
|
WD repeat containing, antisense to TP73 |
| chr1_+_53308398 | 0.33 |
ENST00000371528.1
|
ZYG11A
|
zyg-11 family member A, cell cycle regulator |
| chr1_+_220267429 | 0.32 |
ENST00000366922.1
ENST00000302637.5 |
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr21_-_34185944 | 0.32 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr1_-_3566627 | 0.32 |
ENST00000419924.2
ENST00000270708.7 |
WRAP73
|
WD repeat containing, antisense to TP73 |
| chr1_+_212475148 | 0.30 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr4_-_104119528 | 0.27 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr12_+_53848549 | 0.27 |
ENST00000439930.3
ENST00000548933.1 ENST00000562264.1 |
PCBP2
|
poly(rC) binding protein 2 |
| chr6_+_149887377 | 0.27 |
ENST00000367419.5
|
GINM1
|
glycoprotein integral membrane 1 |
| chr21_-_34185989 | 0.26 |
ENST00000487113.1
ENST00000382373.4 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr21_-_34186006 | 0.26 |
ENST00000490358.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr14_+_56127989 | 0.25 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr20_+_43990576 | 0.23 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr16_+_777118 | 0.23 |
ENST00000562141.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr4_-_104119488 | 0.23 |
ENST00000514974.1
|
CENPE
|
centromere protein E, 312kDa |
| chr5_+_169758393 | 0.21 |
ENST00000521471.1
ENST00000518357.1 ENST00000436248.3 |
CTB-114C7.3
|
CTB-114C7.3 |
| chr15_+_54901540 | 0.20 |
ENST00000539562.2
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr11_+_61891445 | 0.19 |
ENST00000394818.3
ENST00000533896.1 ENST00000278849.4 |
INCENP
|
inner centromere protein antigens 135/155kDa |
| chr3_+_186158169 | 0.18 |
ENST00000435548.1
ENST00000421006.1 |
RP11-78H24.1
|
RP11-78H24.1 |
| chr8_-_74495065 | 0.18 |
ENST00000523533.1
|
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr11_+_110225855 | 0.17 |
ENST00000526605.1
ENST00000526703.1 |
RP11-347E10.1
|
RP11-347E10.1 |
| chr15_+_41057818 | 0.17 |
ENST00000558467.1
|
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr17_-_6524159 | 0.16 |
ENST00000589033.1
|
KIAA0753
|
KIAA0753 |
| chr14_-_68159152 | 0.15 |
ENST00000554035.1
|
RDH11
|
retinol dehydrogenase 11 (all-trans/9-cis/11-cis) |
| chr1_-_149859466 | 0.15 |
ENST00000331128.3
|
HIST2H2AB
|
histone cluster 2, H2ab |
| chr18_-_56985776 | 0.14 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr14_+_56127960 | 0.14 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr17_-_47723943 | 0.11 |
ENST00000510476.1
ENST00000503676.1 |
SPOP
|
speckle-type POZ protein |
| chr22_-_24096562 | 0.11 |
ENST00000398465.3
|
VPREB3
|
pre-B lymphocyte 3 |
| chr3_+_68053359 | 0.10 |
ENST00000478136.1
|
FAM19A1
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 |
| chr17_-_42327236 | 0.09 |
ENST00000399246.2
|
AC003102.1
|
AC003102.1 |
| chrX_+_36254051 | 0.09 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr4_+_158142750 | 0.08 |
ENST00000505888.1
ENST00000449365.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr3_-_27498235 | 0.07 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr2_+_133874577 | 0.07 |
ENST00000596384.1
|
AC011755.1
|
HCG2006742; Protein LOC100996685 |
| chr8_-_67976509 | 0.07 |
ENST00000518747.1
|
COPS5
|
COP9 signalosome subunit 5 |
| chr7_-_55583740 | 0.06 |
ENST00000453256.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr7_-_92855762 | 0.06 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr2_-_183106641 | 0.06 |
ENST00000346717.4
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr2_+_113670548 | 0.06 |
ENST00000263326.3
ENST00000352179.3 ENST00000349806.3 ENST00000353225.3 |
IL37
|
interleukin 37 |
| chr5_+_175490540 | 0.06 |
ENST00000515817.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr2_+_58655520 | 0.05 |
ENST00000455219.3
ENST00000449448.2 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chrX_+_47867194 | 0.05 |
ENST00000376940.3
|
SPACA5
|
sperm acrosome associated 5 |
| chr3_-_139199565 | 0.05 |
ENST00000511956.1
|
RBP2
|
retinol binding protein 2, cellular |
| chr3_-_139199589 | 0.05 |
ENST00000506825.1
|
RBP2
|
retinol binding protein 2, cellular |
| chrX_-_19988382 | 0.05 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chr9_-_37465396 | 0.05 |
ENST00000307750.4
|
ZBTB5
|
zinc finger and BTB domain containing 5 |
| chr12_-_10282836 | 0.05 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr7_+_23210760 | 0.04 |
ENST00000366347.4
|
AC005082.1
|
Uncharacterized protein |
| chr8_+_67976593 | 0.04 |
ENST00000262210.5
ENST00000412460.1 |
CSPP1
|
centrosome and spindle pole associated protein 1 |
| chr18_-_70305745 | 0.04 |
ENST00000581073.1
|
CBLN2
|
cerebellin 2 precursor |
| chrX_+_15767971 | 0.04 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr13_-_41768654 | 0.03 |
ENST00000379483.3
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7 |
| chr4_-_177190364 | 0.03 |
ENST00000296525.3
|
ASB5
|
ankyrin repeat and SOCS box containing 5 |
| chr7_+_871559 | 0.03 |
ENST00000421580.1
|
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr5_-_177207634 | 0.03 |
ENST00000513554.1
ENST00000440605.3 |
FAM153A
|
family with sequence similarity 153, member A |
| chrX_-_15333736 | 0.02 |
ENST00000380470.3
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr2_-_238333919 | 0.02 |
ENST00000409162.1
|
AC112721.1
|
Uncharacterized protein |
| chr1_+_170904612 | 0.02 |
ENST00000367759.4
ENST00000367758.3 |
MROH9
|
maestro heat-like repeat family member 9 |
| chr15_-_51630772 | 0.02 |
ENST00000557858.1
ENST00000558328.1 ENST00000396404.4 ENST00000561075.1 ENST00000405011.2 ENST00000559980.1 ENST00000453807.2 ENST00000396402.1 |
CYP19A1
|
cytochrome P450, family 19, subfamily A, polypeptide 1 |
| chr17_-_29641084 | 0.02 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr19_-_48059113 | 0.02 |
ENST00000391901.3
ENST00000314121.4 ENST00000448976.1 |
ZNF541
|
zinc finger protein 541 |
| chr19_+_50691437 | 0.01 |
ENST00000598205.1
|
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr18_-_46784778 | 0.01 |
ENST00000582399.1
|
DYM
|
dymeclin |
| chr4_+_71458012 | 0.01 |
ENST00000449493.2
|
AMBN
|
ameloblastin (enamel matrix protein) |
| chr4_+_158141899 | 0.00 |
ENST00000264426.9
ENST00000506284.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr4_+_71457970 | 0.00 |
ENST00000322937.6
|
AMBN
|
ameloblastin (enamel matrix protein) |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX6_IRX4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.2 | 1.6 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.1 | 0.8 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.1 | 0.4 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.3 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.5 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 1.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 1.4 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.1 | 0.2 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.1 | 1.3 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 1.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.2 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.3 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 1.0 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.6 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.8 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.2 | 1.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 1.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 1.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.1 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.3 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.8 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.1 | 0.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.2 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.1 | 1.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 1.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.8 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 1.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.3 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |