Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
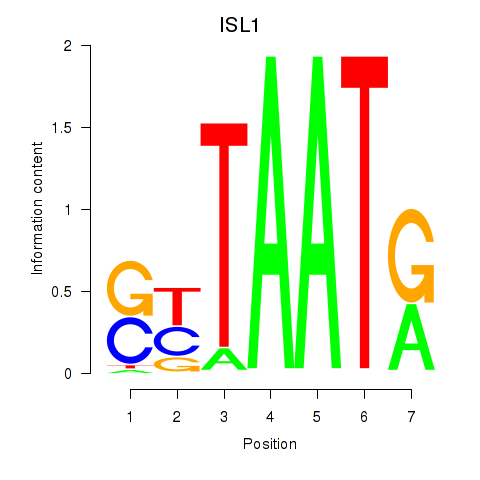
Results for ISL1
Z-value: 0.69
Transcription factors associated with ISL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ISL1
|
ENSG00000016082.10 | ISL LIM homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ISL1 | hg19_v2_chr5_+_50678921_50678921 | -0.27 | 2.5e-01 | Click! |
Activity profile of ISL1 motif
Sorted Z-values of ISL1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_107880508 | 1.31 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
| chr20_+_60174827 | 1.20 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr2_-_175711133 | 1.07 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr7_-_81635106 | 1.04 |
ENST00000443883.1
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chrX_-_30327495 | 0.94 |
ENST00000453287.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr5_+_32710736 | 0.94 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr11_-_115127611 | 0.82 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chr5_+_102455853 | 0.78 |
ENST00000515845.1
ENST00000321521.9 ENST00000507921.1 |
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr6_-_130031358 | 0.78 |
ENST00000368149.2
|
ARHGAP18
|
Rho GTPase activating protein 18 |
| chr6_-_108278456 | 0.77 |
ENST00000429168.1
|
SEC63
|
SEC63 homolog (S. cerevisiae) |
| chr5_-_94417562 | 0.76 |
ENST00000505465.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr11_+_123430948 | 0.75 |
ENST00000529432.1
ENST00000534764.1 |
GRAMD1B
|
GRAM domain containing 1B |
| chrX_+_10031499 | 0.70 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chrX_-_119693745 | 0.69 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr3_+_142342228 | 0.68 |
ENST00000337777.3
|
PLS1
|
plastin 1 |
| chr11_+_7618413 | 0.63 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr12_+_50144381 | 0.62 |
ENST00000552370.1
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr5_+_32711419 | 0.60 |
ENST00000265074.8
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr6_-_131211534 | 0.56 |
ENST00000456097.2
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr5_-_94417339 | 0.56 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr10_+_17270214 | 0.54 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr10_-_95242044 | 0.53 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr9_+_2029019 | 0.53 |
ENST00000382194.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr8_+_26371763 | 0.53 |
ENST00000521913.1
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr4_+_169418195 | 0.53 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr4_+_169418255 | 0.52 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr2_+_183582774 | 0.52 |
ENST00000537515.1
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr7_+_105172612 | 0.51 |
ENST00000493041.1
|
RINT1
|
RAD50 interactor 1 |
| chr8_-_131399110 | 0.50 |
ENST00000521426.1
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr3_-_33686925 | 0.50 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr6_-_111136299 | 0.50 |
ENST00000457688.1
|
CDK19
|
cyclin-dependent kinase 19 |
| chr10_-_115613828 | 0.50 |
ENST00000361384.2
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr15_+_59903975 | 0.50 |
ENST00000560585.1
ENST00000396065.1 |
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr1_-_100598444 | 0.49 |
ENST00000535161.1
ENST00000287482.5 |
SASS6
|
spindle assembly 6 homolog (C. elegans) |
| chr1_-_108231101 | 0.49 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr15_-_72767490 | 0.48 |
ENST00000565181.1
|
RP11-1007O24.3
|
RP11-1007O24.3 |
| chr1_+_200011711 | 0.48 |
ENST00000544748.1
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr1_-_232651312 | 0.48 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr17_-_47045949 | 0.47 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr20_-_25320367 | 0.47 |
ENST00000450393.1
ENST00000491682.1 |
ABHD12
|
abhydrolase domain containing 12 |
| chrX_+_135252050 | 0.46 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chr8_+_53850991 | 0.46 |
ENST00000331251.3
|
NPBWR1
|
neuropeptides B/W receptor 1 |
| chr12_-_102591604 | 0.46 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr5_+_138210919 | 0.45 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr12_+_79439461 | 0.45 |
ENST00000552624.1
|
SYT1
|
synaptotagmin I |
| chr5_-_94417186 | 0.44 |
ENST00000312216.8
ENST00000512425.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr19_-_42759266 | 0.44 |
ENST00000594664.1
|
AC006486.9
|
Uncharacterized protein |
| chr8_-_101321584 | 0.43 |
ENST00000523167.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr10_-_36813162 | 0.43 |
ENST00000440465.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr17_-_57229155 | 0.43 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr12_-_52761262 | 0.43 |
ENST00000257901.3
|
KRT85
|
keratin 85 |
| chr18_-_2982869 | 0.41 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr3_-_138312971 | 0.41 |
ENST00000485115.1
ENST00000484888.1 ENST00000468900.1 ENST00000542237.1 ENST00000481834.1 |
CEP70
|
centrosomal protein 70kDa |
| chr1_-_197169672 | 0.41 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chr17_-_15501932 | 0.40 |
ENST00000583965.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr10_-_95241951 | 0.40 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr5_+_32788945 | 0.40 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chr8_-_19540266 | 0.38 |
ENST00000311540.4
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr11_-_107729504 | 0.38 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr16_+_20775358 | 0.37 |
ENST00000440284.2
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chrX_+_135251835 | 0.37 |
ENST00000456445.1
|
FHL1
|
four and a half LIM domains 1 |
| chr11_-_107729287 | 0.37 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr10_-_27529486 | 0.36 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr21_-_16374688 | 0.36 |
ENST00000411932.1
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr6_+_108977520 | 0.36 |
ENST00000540898.1
|
FOXO3
|
forkhead box O3 |
| chr3_-_27498235 | 0.35 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr3_-_135913298 | 0.35 |
ENST00000434835.2
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chr5_+_102200948 | 0.35 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr5_+_142286887 | 0.35 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr8_-_19540086 | 0.35 |
ENST00000332246.6
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr4_+_4861385 | 0.34 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr2_+_172309634 | 0.34 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr15_+_65822756 | 0.34 |
ENST00000562901.1
ENST00000261875.5 ENST00000442729.2 ENST00000565299.1 ENST00000568793.1 |
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr8_+_21911054 | 0.34 |
ENST00000519850.1
ENST00000381470.3 |
DMTN
|
dematin actin binding protein |
| chr9_-_92112953 | 0.33 |
ENST00000339861.4
ENST00000422704.2 ENST00000455551.2 |
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr6_+_167704798 | 0.33 |
ENST00000230256.3
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr8_+_42873548 | 0.33 |
ENST00000533338.1
ENST00000534420.1 |
HOOK3
RP11-598P20.5
|
hook microtubule-tethering protein 3 Uncharacterized protein |
| chr2_-_208030886 | 0.32 |
ENST00000426163.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr17_+_63096903 | 0.32 |
ENST00000582940.1
|
RP11-160O5.1
|
RP11-160O5.1 |
| chr6_-_111804905 | 0.31 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr22_-_43355858 | 0.31 |
ENST00000402229.1
ENST00000407585.1 ENST00000453079.1 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr2_-_68547061 | 0.30 |
ENST00000263655.3
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr5_-_94417314 | 0.30 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr9_-_86323118 | 0.30 |
ENST00000376395.4
|
UBQLN1
|
ubiquilin 1 |
| chr6_-_24720226 | 0.30 |
ENST00000378102.3
|
C6orf62
|
chromosome 6 open reading frame 62 |
| chr7_-_14026063 | 0.30 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr9_-_134585221 | 0.29 |
ENST00000372190.3
ENST00000427994.1 |
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr12_+_79357815 | 0.29 |
ENST00000547046.1
|
SYT1
|
synaptotagmin I |
| chr2_-_175711978 | 0.29 |
ENST00000409089.2
|
CHN1
|
chimerin 1 |
| chr12_-_96793142 | 0.29 |
ENST00000552262.1
ENST00000551816.1 ENST00000552496.1 |
CDK17
|
cyclin-dependent kinase 17 |
| chr16_+_20775024 | 0.29 |
ENST00000289416.5
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr6_+_167704838 | 0.28 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr5_+_34757309 | 0.28 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr5_-_102455801 | 0.28 |
ENST00000508629.1
ENST00000399004.2 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chr6_+_35996859 | 0.28 |
ENST00000472333.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr19_-_42806842 | 0.27 |
ENST00000596265.1
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr13_-_99667960 | 0.27 |
ENST00000448493.2
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr15_+_59279851 | 0.27 |
ENST00000348370.4
ENST00000434298.1 ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr17_-_40729681 | 0.26 |
ENST00000590760.1
ENST00000587209.1 ENST00000393795.3 ENST00000253789.5 |
PSMC3IP
|
PSMC3 interacting protein |
| chr7_-_150675372 | 0.26 |
ENST00000262186.5
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr10_-_89577910 | 0.26 |
ENST00000308448.7
ENST00000541004.1 |
ATAD1
|
ATPase family, AAA domain containing 1 |
| chrX_+_135251783 | 0.26 |
ENST00000394153.2
|
FHL1
|
four and a half LIM domains 1 |
| chr8_+_35092959 | 0.25 |
ENST00000404895.2
|
UNC5D
|
unc-5 homolog D (C. elegans) |
| chr18_-_19283649 | 0.25 |
ENST00000584464.1
ENST00000578270.1 |
ABHD3
|
abhydrolase domain containing 3 |
| chr17_+_66521936 | 0.25 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr13_+_78315466 | 0.24 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chrX_+_72783026 | 0.24 |
ENST00000373504.6
ENST00000373502.5 |
CHIC1
|
cysteine-rich hydrophobic domain 1 |
| chr8_+_19536083 | 0.24 |
ENST00000519803.1
|
RP11-1105O14.1
|
RP11-1105O14.1 |
| chr1_-_8000872 | 0.24 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chrX_+_114874727 | 0.24 |
ENST00000543070.1
|
PLS3
|
plastin 3 |
| chr5_-_79950775 | 0.24 |
ENST00000439211.2
|
DHFR
|
dihydrofolate reductase |
| chr10_-_17659357 | 0.24 |
ENST00000326961.6
ENST00000361271.3 |
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr20_+_19870167 | 0.23 |
ENST00000440354.2
|
RIN2
|
Ras and Rab interactor 2 |
| chr6_-_111136513 | 0.23 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr3_-_33686743 | 0.23 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr6_+_144904334 | 0.22 |
ENST00000367526.4
|
UTRN
|
utrophin |
| chr5_+_134181755 | 0.22 |
ENST00000504727.1
ENST00000435259.2 ENST00000508791.1 |
C5orf24
|
chromosome 5 open reading frame 24 |
| chr12_+_54410664 | 0.22 |
ENST00000303406.4
|
HOXC4
|
homeobox C4 |
| chr17_-_64225508 | 0.22 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr4_-_128887069 | 0.22 |
ENST00000541133.1
ENST00000296468.3 ENST00000513559.1 |
MFSD8
|
major facilitator superfamily domain containing 8 |
| chr9_-_96108696 | 0.22 |
ENST00000375419.1
|
C9orf129
|
chromosome 9 open reading frame 129 |
| chr3_+_158787041 | 0.21 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr14_+_62164340 | 0.21 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr13_+_108870714 | 0.21 |
ENST00000375898.3
|
ABHD13
|
abhydrolase domain containing 13 |
| chr9_-_113761720 | 0.21 |
ENST00000541779.1
ENST00000374430.2 |
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr9_+_128510454 | 0.21 |
ENST00000491787.3
ENST00000447726.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr11_-_66496430 | 0.21 |
ENST00000533211.1
|
SPTBN2
|
spectrin, beta, non-erythrocytic 2 |
| chr4_+_113970772 | 0.20 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr12_-_121454148 | 0.20 |
ENST00000535367.1
ENST00000538296.1 ENST00000445832.3 ENST00000536407.2 ENST00000366211.2 ENST00000539736.1 ENST00000288757.3 ENST00000537817.1 |
C12orf43
|
chromosome 12 open reading frame 43 |
| chr5_+_140248518 | 0.20 |
ENST00000398640.2
|
PCDHA11
|
protocadherin alpha 11 |
| chr10_-_116444371 | 0.20 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr5_+_135468516 | 0.19 |
ENST00000507118.1
ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5
|
SMAD family member 5 |
| chr8_-_101571964 | 0.19 |
ENST00000520552.1
ENST00000521345.1 ENST00000523000.1 ENST00000335659.3 ENST00000358990.3 ENST00000519597.1 |
ANKRD46
|
ankyrin repeat domain 46 |
| chr14_+_61449076 | 0.19 |
ENST00000526105.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr15_-_55541227 | 0.19 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_+_15986364 | 0.18 |
ENST00000345034.1
|
RSC1A1
|
regulatory solute carrier protein, family 1, member 1 |
| chr16_-_3350614 | 0.18 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr1_-_119682812 | 0.18 |
ENST00000537870.1
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr12_+_14572070 | 0.18 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr17_-_2615031 | 0.17 |
ENST00000576885.1
ENST00000574426.2 |
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr13_+_78315295 | 0.17 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr13_+_28527647 | 0.17 |
ENST00000567234.1
|
LINC00543
|
long intergenic non-protein coding RNA 543 |
| chrX_-_15683147 | 0.16 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr2_-_224467002 | 0.16 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr2_-_175202151 | 0.16 |
ENST00000595354.1
|
AC018470.1
|
Uncharacterized protein FLJ46347 |
| chr6_+_149539053 | 0.16 |
ENST00000451095.1
|
RP1-111D6.3
|
RP1-111D6.3 |
| chr7_+_90012986 | 0.16 |
ENST00000416322.1
|
CLDN12
|
claudin 12 |
| chr3_-_149095652 | 0.15 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr4_+_139694701 | 0.15 |
ENST00000502606.1
|
RP11-98O2.1
|
RP11-98O2.1 |
| chr7_-_35734176 | 0.15 |
ENST00000413517.1
ENST00000438224.1 |
HERPUD2
|
HERPUD family member 2 |
| chr17_-_39341594 | 0.15 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr12_+_9066472 | 0.15 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr2_-_208030295 | 0.15 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr17_+_26698677 | 0.15 |
ENST00000457710.3
|
SARM1
|
sterile alpha and TIR motif containing 1 |
| chr2_-_55920952 | 0.15 |
ENST00000447944.2
|
PNPT1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr3_+_35721106 | 0.14 |
ENST00000474696.1
ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr2_-_208030647 | 0.14 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr13_+_78315348 | 0.14 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr2_-_74735707 | 0.14 |
ENST00000233630.6
|
PCGF1
|
polycomb group ring finger 1 |
| chr10_+_51572408 | 0.13 |
ENST00000374082.1
|
NCOA4
|
nuclear receptor coactivator 4 |
| chr1_+_221054411 | 0.13 |
ENST00000427693.1
|
HLX
|
H2.0-like homeobox |
| chr3_+_157154578 | 0.13 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr9_+_131084846 | 0.13 |
ENST00000608951.1
|
COQ4
|
coenzyme Q4 |
| chr7_+_13141010 | 0.12 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr10_+_114710425 | 0.12 |
ENST00000352065.5
ENST00000369395.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr14_+_20811766 | 0.12 |
ENST00000250416.5
ENST00000527915.1 |
PARP2
|
poly (ADP-ribose) polymerase 2 |
| chr1_-_93257951 | 0.11 |
ENST00000543509.1
ENST00000370331.1 ENST00000540033.1 |
EVI5
|
ecotropic viral integration site 5 |
| chr12_+_54402790 | 0.11 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr1_+_150980889 | 0.11 |
ENST00000450884.1
ENST00000271620.3 ENST00000271619.8 ENST00000368937.1 ENST00000431193.1 ENST00000368936.1 |
PRUNE
|
prune exopolyphosphatase |
| chr6_+_15401075 | 0.11 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr13_+_111893533 | 0.11 |
ENST00000478679.1
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr17_-_41132010 | 0.10 |
ENST00000409103.1
ENST00000360221.4 |
PTGES3L-AARSD1
|
PTGES3L-AARSD1 readthrough |
| chrX_-_102941596 | 0.09 |
ENST00000441076.2
ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr14_-_77495007 | 0.09 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr2_+_149402009 | 0.09 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr7_+_16793160 | 0.09 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chrX_-_102983417 | 0.08 |
ENST00000372617.4
|
GLRA4
|
glycine receptor, alpha 4 |
| chr3_-_49066811 | 0.08 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr2_-_224467093 | 0.08 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr1_+_12776118 | 0.08 |
ENST00000332530.3
ENST00000359318.5 |
AADACL3
|
arylacetamide deacetylase-like 3 |
| chr2_+_28618532 | 0.08 |
ENST00000545753.1
|
FOSL2
|
FOS-like antigen 2 |
| chr11_-_40315640 | 0.08 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr8_-_79717750 | 0.08 |
ENST00000263851.4
ENST00000379113.2 |
IL7
|
interleukin 7 |
| chr5_-_142782862 | 0.07 |
ENST00000415690.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr1_-_169555709 | 0.07 |
ENST00000546081.1
|
F5
|
coagulation factor V (proaccelerin, labile factor) |
| chr3_-_100712352 | 0.07 |
ENST00000471714.1
ENST00000284322.5 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr1_+_161736072 | 0.07 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr1_+_100598691 | 0.07 |
ENST00000370143.1
ENST00000370141.2 |
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr18_-_21977748 | 0.07 |
ENST00000399441.4
ENST00000319481.3 |
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr11_-_7904464 | 0.07 |
ENST00000502624.2
ENST00000527565.1 |
RP11-35J10.5
|
RP11-35J10.5 |
| chr1_+_109265033 | 0.07 |
ENST00000445274.1
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr1_+_100598742 | 0.06 |
ENST00000370139.1
|
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr16_+_47496023 | 0.06 |
ENST00000567200.1
|
PHKB
|
phosphorylase kinase, beta |
| chr2_+_172543919 | 0.06 |
ENST00000452242.1
ENST00000340296.4 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr1_+_217804661 | 0.06 |
ENST00000366933.4
|
SPATA17
|
spermatogenesis associated 17 |
| chr2_+_128377550 | 0.06 |
ENST00000437387.1
ENST00000409090.1 |
MYO7B
|
myosin VIIB |
| chr22_-_32651326 | 0.06 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr9_+_134165195 | 0.06 |
ENST00000372261.1
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3 |
| chr5_-_56778635 | 0.06 |
ENST00000423391.1
|
ACTBL2
|
actin, beta-like 2 |
| chr12_+_116955659 | 0.06 |
ENST00000552992.1
|
RP11-148B3.1
|
RP11-148B3.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ISL1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.8 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 1.0 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.1 | 0.7 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.5 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.1 | 0.4 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.4 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.5 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.3 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.1 | 0.3 | GO:1901340 | negative regulation of store-operated calcium channel activity(GO:1901340) |
| 0.1 | 0.7 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.7 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 0.5 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 1.5 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 0.6 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.1 | 0.3 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.5 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.1 | 0.5 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 1.3 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.9 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.2 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.1 | 0.5 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.5 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.1 | 0.2 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.1 | 0.4 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 0.3 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.1 | 0.3 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.2 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.1 | 0.7 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:2000627 | rRNA import into mitochondrion(GO:0035928) regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.0 | 0.2 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.9 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.7 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.5 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 1.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.5 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.3 | GO:1901731 | positive regulation of platelet aggregation(GO:1901731) |
| 0.0 | 0.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.7 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.5 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.3 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 1.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.6 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0044793 | negative regulation by host of viral process(GO:0044793) negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.3 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) |
| 0.0 | 0.2 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 1.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.0 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 1.1 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.1 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.4 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 1.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.7 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.5 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.7 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.3 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 1.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.7 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.9 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.5 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0005902 | microvillus(GO:0005902) actin-based cell projection(GO:0098858) |
| 0.0 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 1.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.2 | GO:0070938 | contractile ring(GO:0070938) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.2 | 1.5 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 0.5 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.1 | 0.5 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.7 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.1 | 0.8 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.6 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 0.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.7 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.4 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.3 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 1.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.5 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 1.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.9 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 1.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.5 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 1.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.0 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.7 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 1.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 3.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |