Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
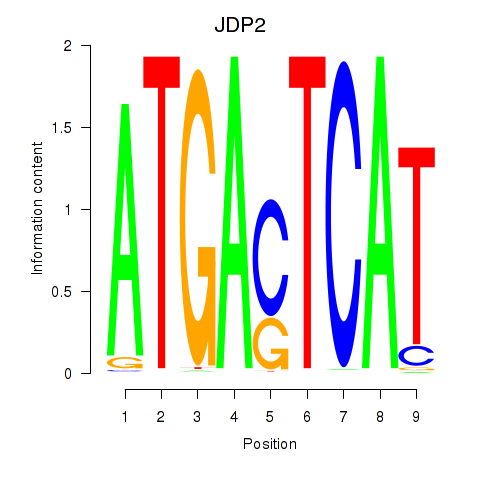
Results for JDP2
Z-value: 2.15
Transcription factors associated with JDP2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JDP2
|
ENSG00000140044.8 | Jun dimerization protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| JDP2 | hg19_v2_chr14_+_75894391_75894435 | 0.37 | 1.0e-01 | Click! |
Activity profile of JDP2 motif
Sorted Z-values of JDP2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_56056888 | 11.69 |
ENST00000592464.1
ENST00000420723.3 |
SBK3
|
SH3 domain binding kinase family, member 3 |
| chr11_-_102401469 | 10.77 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr20_-_62129163 | 9.50 |
ENST00000298049.7
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr8_-_27472198 | 7.75 |
ENST00000519472.1
ENST00000523589.1 ENST00000522413.1 ENST00000523396.1 ENST00000560366.1 |
CLU
|
clusterin |
| chrY_+_15016725 | 7.64 |
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr20_+_43029911 | 7.39 |
ENST00000443598.2
ENST00000316099.4 ENST00000415691.2 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr9_+_118950325 | 7.28 |
ENST00000534838.1
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr1_+_156084461 | 6.85 |
ENST00000347559.2
ENST00000361308.4 ENST00000368300.4 ENST00000368299.3 |
LMNA
|
lamin A/C |
| chrX_-_15288154 | 6.84 |
ENST00000380483.3
ENST00000380485.3 ENST00000380488.4 |
ASB9
|
ankyrin repeat and SOCS box containing 9 |
| chr17_+_4853442 | 6.77 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr16_-_57798253 | 6.58 |
ENST00000565270.1
|
KIFC3
|
kinesin family member C3 |
| chr16_-_69760409 | 6.29 |
ENST00000561500.1
ENST00000439109.2 ENST00000564043.1 ENST00000379046.2 ENST00000379047.3 |
NQO1
|
NAD(P)H dehydrogenase, quinone 1 |
| chr10_+_17270214 | 6.25 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr5_-_75919253 | 6.25 |
ENST00000296641.4
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr20_-_17539456 | 6.10 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chr17_+_48712206 | 5.96 |
ENST00000427699.1
ENST00000285238.8 |
ABCC3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr7_+_30960915 | 5.89 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr9_-_123639304 | 5.56 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19 |
| chr1_-_153521597 | 5.56 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr12_+_52695617 | 5.53 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr2_+_87754989 | 5.49 |
ENST00000409898.2
ENST00000419680.2 ENST00000414584.1 ENST00000455131.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr5_-_90610200 | 5.49 |
ENST00000511918.1
ENST00000513626.1 ENST00000607854.1 |
LUCAT1
RP11-213H15.4
|
lung cancer associated transcript 1 (non-protein coding) RP11-213H15.4 |
| chr12_-_53343602 | 5.44 |
ENST00000546897.1
ENST00000552551.1 |
KRT8
|
keratin 8 |
| chr12_-_53343560 | 5.33 |
ENST00000548998.1
|
KRT8
|
keratin 8 |
| chr17_+_48712138 | 4.96 |
ENST00000515707.1
|
ABCC3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr5_-_75919217 | 4.95 |
ENST00000504899.1
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr16_-_30997533 | 4.95 |
ENST00000602217.1
|
AC135048.1
|
Uncharacterized protein |
| chr4_+_84457529 | 4.75 |
ENST00000264409.4
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr11_-_2924720 | 4.75 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr4_+_84457250 | 4.71 |
ENST00000395226.2
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr12_-_53343633 | 4.69 |
ENST00000546826.1
|
KRT8
|
keratin 8 |
| chr1_-_153521714 | 4.47 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr1_+_156096336 | 4.45 |
ENST00000504687.1
ENST00000473598.2 |
LMNA
|
lamin A/C |
| chr9_-_123638633 | 4.44 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19 |
| chr3_+_48507210 | 4.40 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr17_-_27045427 | 4.32 |
ENST00000301043.6
ENST00000412625.1 |
RAB34
|
RAB34, member RAS oncogene family |
| chr2_+_152214098 | 4.30 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr13_-_67802549 | 4.26 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr12_-_102591604 | 4.17 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr16_+_15596123 | 4.16 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr2_-_175712270 | 4.13 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr3_+_98699880 | 3.84 |
ENST00000473756.1
|
LINC00973
|
long intergenic non-protein coding RNA 973 |
| chr11_+_58874701 | 3.79 |
ENST00000529618.1
ENST00000534403.1 ENST00000343597.3 |
FAM111B
|
family with sequence similarity 111, member B |
| chr17_-_27045165 | 3.75 |
ENST00000436730.3
ENST00000450529.1 ENST00000583538.1 ENST00000419712.3 ENST00000580843.2 ENST00000582934.1 ENST00000415040.2 ENST00000353676.5 ENST00000453384.3 ENST00000447716.1 |
RAB34
|
RAB34, member RAS oncogene family |
| chr7_-_47492782 | 3.69 |
ENST00000413551.1
|
TNS3
|
tensin 3 |
| chr12_-_105352047 | 3.67 |
ENST00000432951.1
ENST00000415674.1 ENST00000424946.1 |
SLC41A2
|
solute carrier family 41 (magnesium transporter), member 2 |
| chr3_+_48507621 | 3.64 |
ENST00000456089.1
|
TREX1
|
three prime repair exonuclease 1 |
| chr17_-_27044903 | 3.60 |
ENST00000395245.3
|
RAB34
|
RAB34, member RAS oncogene family |
| chr19_-_46285646 | 3.58 |
ENST00000458663.2
|
DMPK
|
dystrophia myotonica-protein kinase |
| chr2_+_10262442 | 3.55 |
ENST00000360566.2
|
RRM2
|
ribonucleotide reductase M2 |
| chr17_+_25799008 | 3.55 |
ENST00000583370.1
ENST00000398988.3 ENST00000268763.6 |
KSR1
|
kinase suppressor of ras 1 |
| chr10_+_88854926 | 3.42 |
ENST00000298784.1
ENST00000298786.4 |
FAM35A
|
family with sequence similarity 35, member A |
| chr15_-_54025300 | 3.41 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr3_+_69134124 | 3.39 |
ENST00000478935.1
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr3_+_28283069 | 3.38 |
ENST00000466830.1
ENST00000423894.1 |
CMC1
|
C-x(9)-C motif containing 1 |
| chr12_+_7023491 | 3.38 |
ENST00000541477.1
ENST00000229277.1 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr10_+_94594351 | 3.31 |
ENST00000371552.4
|
EXOC6
|
exocyst complex component 6 |
| chr12_+_7023735 | 3.27 |
ENST00000538763.1
ENST00000544774.1 ENST00000545045.2 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr17_-_27044810 | 3.24 |
ENST00000395242.2
|
RAB34
|
RAB34, member RAS oncogene family |
| chr7_-_155604967 | 3.23 |
ENST00000297261.2
|
SHH
|
sonic hedgehog |
| chr12_-_6960407 | 3.18 |
ENST00000540683.1
ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3
|
cell division cycle associated 3 |
| chr2_+_87754887 | 3.10 |
ENST00000409054.1
ENST00000331944.6 ENST00000409139.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr1_-_244006528 | 3.07 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr2_-_145275228 | 3.05 |
ENST00000427902.1
ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr11_+_67250490 | 2.99 |
ENST00000528641.2
ENST00000279146.3 |
AIP
|
aryl hydrocarbon receptor interacting protein |
| chr2_+_87755054 | 2.89 |
ENST00000423846.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr2_+_46926048 | 2.83 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr17_-_70417365 | 2.83 |
ENST00000580948.1
|
LINC00511
|
long intergenic non-protein coding RNA 511 |
| chr2_+_202122826 | 2.82 |
ENST00000413726.1
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr2_-_175711978 | 2.80 |
ENST00000409089.2
|
CHN1
|
chimerin 1 |
| chr12_-_53297432 | 2.78 |
ENST00000546900.1
|
KRT8
|
keratin 8 |
| chr5_+_72143988 | 2.74 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr11_+_58874658 | 2.73 |
ENST00000411426.1
|
FAM111B
|
family with sequence similarity 111, member B |
| chr17_-_27045405 | 2.73 |
ENST00000430132.2
|
RAB34
|
RAB34, member RAS oncogene family |
| chr11_-_2950642 | 2.72 |
ENST00000314222.4
|
PHLDA2
|
pleckstrin homology-like domain, family A, member 2 |
| chr1_+_33722080 | 2.69 |
ENST00000483388.1
ENST00000539719.1 |
ZNF362
|
zinc finger protein 362 |
| chr22_-_43398442 | 2.61 |
ENST00000422336.1
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr19_-_51872233 | 2.59 |
ENST00000601435.1
ENST00000291715.1 |
CLDND2
|
claudin domain containing 2 |
| chr2_-_175712479 | 2.57 |
ENST00000443238.1
|
CHN1
|
chimerin 1 |
| chr16_-_31161380 | 2.55 |
ENST00000569305.1
ENST00000418068.2 ENST00000268281.4 |
PRSS36
|
protease, serine, 36 |
| chr8_+_40018977 | 2.45 |
ENST00000520487.1
|
RP11-470M17.2
|
RP11-470M17.2 |
| chr14_-_51562037 | 2.42 |
ENST00000338969.5
|
TRIM9
|
tripartite motif containing 9 |
| chr14_-_51561784 | 2.33 |
ENST00000360392.4
|
TRIM9
|
tripartite motif containing 9 |
| chr17_-_47785504 | 2.32 |
ENST00000514907.1
ENST00000503334.1 ENST00000508520.1 |
SLC35B1
|
solute carrier family 35, member B1 |
| chr15_+_89182156 | 2.30 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr4_-_987217 | 2.24 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr2_-_145275109 | 2.21 |
ENST00000431672.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_11606275 | 2.19 |
ENST00000381525.3
ENST00000362009.4 |
E2F6
|
E2F transcription factor 6 |
| chr8_-_123139423 | 2.19 |
ENST00000523792.1
|
RP11-398G24.2
|
RP11-398G24.2 |
| chr5_+_169758393 | 2.19 |
ENST00000521471.1
ENST00000518357.1 ENST00000436248.3 |
CTB-114C7.3
|
CTB-114C7.3 |
| chr17_+_80317121 | 2.19 |
ENST00000333437.4
|
TEX19
|
testis expressed 19 |
| chr3_-_149293990 | 2.18 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr2_+_7073174 | 2.14 |
ENST00000416587.1
|
RNF144A
|
ring finger protein 144A |
| chrX_+_153770421 | 2.14 |
ENST00000369609.5
ENST00000369607.1 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr16_+_50300427 | 2.12 |
ENST00000394697.2
ENST00000566433.2 ENST00000538642.1 |
ADCY7
|
adenylate cyclase 7 |
| chr15_+_65843130 | 2.06 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr11_-_2924970 | 2.01 |
ENST00000533594.1
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr17_-_47786375 | 1.99 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr11_+_122526383 | 1.96 |
ENST00000284273.5
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B |
| chr14_-_23426231 | 1.92 |
ENST00000556915.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr2_+_202122703 | 1.88 |
ENST00000447616.1
ENST00000358485.4 |
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr2_+_27505260 | 1.87 |
ENST00000380075.2
ENST00000296098.4 |
TRIM54
|
tripartite motif containing 54 |
| chr15_+_75335604 | 1.87 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chrX_-_154493791 | 1.84 |
ENST00000369454.3
|
RAB39B
|
RAB39B, member RAS oncogene family |
| chr11_-_75201791 | 1.80 |
ENST00000529721.1
|
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr15_+_89182178 | 1.78 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr2_-_121624973 | 1.77 |
ENST00000603720.1
|
RP11-297J22.1
|
RP11-297J22.1 |
| chr2_+_69201705 | 1.74 |
ENST00000377938.2
|
GKN1
|
gastrokine 1 |
| chr16_+_2083265 | 1.73 |
ENST00000565855.1
ENST00000566198.1 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr11_+_20620946 | 1.71 |
ENST00000525748.1
|
SLC6A5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chrX_-_19689106 | 1.66 |
ENST00000379716.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr8_-_38386175 | 1.60 |
ENST00000437935.2
ENST00000358138.1 |
C8orf86
|
chromosome 8 open reading frame 86 |
| chr11_-_10920838 | 1.59 |
ENST00000503469.2
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr17_-_53800217 | 1.58 |
ENST00000424486.2
|
TMEM100
|
transmembrane protein 100 |
| chr17_+_76311791 | 1.55 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr15_+_89181974 | 1.55 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr5_+_179247759 | 1.55 |
ENST00000389805.4
ENST00000504627.1 ENST00000402874.3 ENST00000510187.1 |
SQSTM1
|
sequestosome 1 |
| chr1_-_246580705 | 1.52 |
ENST00000541742.1
|
SMYD3
|
SET and MYND domain containing 3 |
| chr11_+_82783097 | 1.50 |
ENST00000501011.2
ENST00000527627.1 ENST00000526795.1 ENST00000533528.1 ENST00000533708.1 ENST00000534499.1 |
RAB30-AS1
|
RAB30 antisense RNA 1 (head to head) |
| chr6_-_31745085 | 1.49 |
ENST00000375686.3
ENST00000447450.1 |
VWA7
|
von Willebrand factor A domain containing 7 |
| chr5_+_135394840 | 1.48 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr2_-_224467002 | 1.48 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr9_+_123906331 | 1.45 |
ENST00000431571.1
|
CNTRL
|
centriolin |
| chr3_-_191000172 | 1.45 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr10_+_104155450 | 1.43 |
ENST00000471698.1
ENST00000189444.6 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr3_-_45957088 | 1.41 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr16_+_31366536 | 1.40 |
ENST00000562522.1
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr17_-_1553346 | 1.40 |
ENST00000301336.6
|
RILP
|
Rab interacting lysosomal protein |
| chr19_-_56047661 | 1.39 |
ENST00000344158.3
|
SBK2
|
SH3 domain binding kinase family, member 2 |
| chr12_-_56101647 | 1.38 |
ENST00000347027.6
ENST00000257879.6 ENST00000257880.7 ENST00000394230.2 ENST00000394229.2 |
ITGA7
|
integrin, alpha 7 |
| chr7_-_135433460 | 1.38 |
ENST00000415751.1
|
FAM180A
|
family with sequence similarity 180, member A |
| chr14_+_32476072 | 1.36 |
ENST00000556949.1
|
RP11-187E13.2
|
Uncharacterized protein |
| chr17_-_8263538 | 1.35 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr3_-_45957534 | 1.35 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr19_+_38880695 | 1.35 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr14_+_73525265 | 1.32 |
ENST00000525161.1
|
RBM25
|
RNA binding motif protein 25 |
| chr6_-_31745037 | 1.30 |
ENST00000375688.4
|
VWA7
|
von Willebrand factor A domain containing 7 |
| chr14_+_38065052 | 1.27 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr1_+_154378049 | 1.26 |
ENST00000512471.1
|
IL6R
|
interleukin 6 receptor |
| chr4_+_120056939 | 1.26 |
ENST00000307128.5
|
MYOZ2
|
myozenin 2 |
| chr5_-_94417562 | 1.26 |
ENST00000505465.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr18_+_2846972 | 1.24 |
ENST00000254528.3
|
EMILIN2
|
elastin microfibril interfacer 2 |
| chr20_-_48782639 | 1.24 |
ENST00000435301.2
|
RP11-112L6.3
|
RP11-112L6.3 |
| chr5_-_67730240 | 1.24 |
ENST00000507733.1
|
CTC-537E7.3
|
CTC-537E7.3 |
| chr2_-_11605966 | 1.24 |
ENST00000307236.4
ENST00000542100.1 ENST00000546212.1 |
E2F6
|
E2F transcription factor 6 |
| chr10_-_31288398 | 1.24 |
ENST00000538351.2
|
ZNF438
|
zinc finger protein 438 |
| chr6_-_84140757 | 1.23 |
ENST00000541327.1
ENST00000369705.3 ENST00000543031.1 |
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr19_-_5903714 | 1.22 |
ENST00000586349.1
ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12
NDUFA11
FUT5
|
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr17_-_55162360 | 1.22 |
ENST00000576871.1
ENST00000576313.1 |
RP11-166P13.3
|
RP11-166P13.3 |
| chr10_-_21463116 | 1.22 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chr19_-_45927622 | 1.21 |
ENST00000300853.3
ENST00000589165.1 |
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr3_+_57094469 | 1.21 |
ENST00000334325.1
|
SPATA12
|
spermatogenesis associated 12 |
| chr3_-_122512619 | 1.20 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr2_-_65593784 | 1.18 |
ENST00000443619.2
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr17_+_55162453 | 1.18 |
ENST00000575322.1
ENST00000337714.3 ENST00000314126.3 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr17_-_38911580 | 1.15 |
ENST00000312150.4
|
KRT25
|
keratin 25 |
| chr9_-_34662651 | 1.14 |
ENST00000259631.4
|
CCL27
|
chemokine (C-C motif) ligand 27 |
| chr17_-_73775839 | 1.13 |
ENST00000592643.1
ENST00000591890.1 ENST00000587171.1 ENST00000254810.4 ENST00000589599.1 |
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr11_+_57308979 | 1.12 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr13_-_36429763 | 1.09 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr7_-_135433534 | 1.09 |
ENST00000338588.3
|
FAM180A
|
family with sequence similarity 180, member A |
| chr14_-_54317532 | 1.08 |
ENST00000418927.1
|
AL162759.1
|
AL162759.1 |
| chr17_+_79650962 | 1.07 |
ENST00000329138.4
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr14_+_103800513 | 1.06 |
ENST00000560338.1
ENST00000560763.1 ENST00000558316.1 ENST00000558265.1 |
EIF5
|
eukaryotic translation initiation factor 5 |
| chr11_-_10920714 | 1.05 |
ENST00000533941.1
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr12_+_48152774 | 1.05 |
ENST00000549243.1
|
SLC48A1
|
solute carrier family 48 (heme transporter), member 1 |
| chr16_-_75301886 | 1.05 |
ENST00000393422.2
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr17_+_75315654 | 1.04 |
ENST00000590595.1
|
SEPT9
|
septin 9 |
| chr17_-_47287928 | 1.04 |
ENST00000507680.1
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr14_+_103801140 | 1.04 |
ENST00000561325.1
ENST00000392715.2 ENST00000559130.1 ENST00000559532.1 ENST00000558506.1 |
EIF5
|
eukaryotic translation initiation factor 5 |
| chr12_-_719573 | 1.03 |
ENST00000397265.3
|
NINJ2
|
ninjurin 2 |
| chr1_-_235116495 | 1.03 |
ENST00000549744.1
|
RP11-443B7.3
|
RP11-443B7.3 |
| chr17_+_47287749 | 1.02 |
ENST00000419580.2
|
ABI3
|
ABI family, member 3 |
| chrX_-_20074895 | 1.01 |
ENST00000543767.1
|
MAP7D2
|
MAP7 domain containing 2 |
| chr15_+_57211318 | 1.01 |
ENST00000557947.1
|
TCF12
|
transcription factor 12 |
| chr2_-_145275211 | 0.99 |
ENST00000462355.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr13_+_53602894 | 0.99 |
ENST00000219022.2
|
OLFM4
|
olfactomedin 4 |
| chr15_-_60690163 | 0.99 |
ENST00000558998.1
ENST00000560165.1 ENST00000557986.1 ENST00000559780.1 ENST00000559467.1 ENST00000559956.1 ENST00000332680.4 ENST00000396024.3 ENST00000421017.2 ENST00000560466.1 ENST00000558132.1 ENST00000559113.1 ENST00000557906.1 ENST00000558558.1 ENST00000560468.1 ENST00000559370.1 ENST00000558169.1 ENST00000559725.1 ENST00000558985.1 ENST00000451270.2 |
ANXA2
|
annexin A2 |
| chr12_+_113860042 | 0.99 |
ENST00000403593.4
|
SDSL
|
serine dehydratase-like |
| chr12_+_113860160 | 0.93 |
ENST00000553248.1
ENST00000345635.4 ENST00000547802.1 |
SDSL
|
serine dehydratase-like |
| chrX_-_48901012 | 0.93 |
ENST00000315869.7
|
TFE3
|
transcription factor binding to IGHM enhancer 3 |
| chr16_+_30211181 | 0.92 |
ENST00000395138.2
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr16_+_31366455 | 0.91 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr11_-_84028339 | 0.88 |
ENST00000398301.2
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr10_-_105238997 | 0.87 |
ENST00000369783.4
|
CALHM3
|
calcium homeostasis modulator 3 |
| chr9_+_140135665 | 0.86 |
ENST00000340384.4
|
TUBB4B
|
tubulin, beta 4B class IVb |
| chr11_-_57417367 | 0.85 |
ENST00000534810.1
|
YPEL4
|
yippee-like 4 (Drosophila) |
| chr1_-_160231451 | 0.85 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr9_-_91793675 | 0.84 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr17_-_39156138 | 0.83 |
ENST00000391587.1
|
KRTAP3-2
|
keratin associated protein 3-2 |
| chr11_-_57417405 | 0.82 |
ENST00000524669.1
ENST00000300022.3 |
YPEL4
|
yippee-like 4 (Drosophila) |
| chr6_+_63921351 | 0.81 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr7_+_872107 | 0.81 |
ENST00000405266.1
ENST00000401592.1 ENST00000403868.1 ENST00000425407.2 |
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr2_-_220173685 | 0.79 |
ENST00000423636.2
ENST00000442029.1 ENST00000412847.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr11_-_84028180 | 0.79 |
ENST00000280241.8
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr17_+_66245341 | 0.78 |
ENST00000577985.1
|
AMZ2
|
archaelysin family metallopeptidase 2 |
| chr6_+_122720681 | 0.78 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr12_-_105352080 | 0.77 |
ENST00000433540.1
|
SLC41A2
|
solute carrier family 41 (magnesium transporter), member 2 |
| chr11_-_68780824 | 0.76 |
ENST00000441623.1
ENST00000309099.6 |
MRGPRF
|
MAS-related GPR, member F |
| chr6_-_31704282 | 0.75 |
ENST00000375784.3
ENST00000375779.2 |
CLIC1
|
chloride intracellular channel 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of JDP2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 2.0 | 5.9 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 1.8 | 10.9 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 1.2 | 4.8 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 1.2 | 17.6 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 1.1 | 5.6 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.1 | 4.3 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 1.1 | 3.2 | GO:0060738 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of immature T cell proliferation in thymus(GO:0033092) right lung development(GO:0060458) primary prostatic bud elongation(GO:0060516) epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 1.1 | 18.2 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 1.0 | 7.8 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.9 | 4.7 | GO:0052053 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.9 | 2.8 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.9 | 2.7 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.8 | 12.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.8 | 10.0 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.7 | 7.4 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.7 | 6.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.7 | 6.6 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.5 | 11.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.5 | 4.2 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.5 | 10.8 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.5 | 1.4 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.4 | 9.8 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.4 | 4.2 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.4 | 1.1 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.4 | 1.4 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.3 | 3.0 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.3 | 1.2 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.3 | 2.7 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.3 | 3.6 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.2 | 1.0 | GO:0052229 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.2 | 7.3 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.2 | 1.8 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.2 | 2.2 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 2.6 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.2 | 13.7 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 1.2 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.2 | 0.8 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.2 | 0.6 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.2 | 9.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.2 | 1.7 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.2 | 3.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 3.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 2.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.2 | 0.5 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.2 | 3.4 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.2 | 2.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 7.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.7 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.1 | 4.3 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 6.3 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.1 | 8.0 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 1.9 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.1 | 1.6 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 1.5 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.1 | 0.9 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 0.6 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 2.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.6 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.1 | 1.0 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.1 | 7.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.0 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 2.0 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.1 | 2.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 1.2 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 0.8 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 0.2 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
| 0.1 | 1.9 | GO:0034030 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.1 | 1.0 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.1 | 0.6 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.1 | 1.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 3.9 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.1 | 1.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 0.6 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 1.3 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.2 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.2 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.3 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.9 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.0 | 0.7 | GO:1990182 | exosomal secretion(GO:1990182) |
| 0.0 | 1.9 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 4.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 3.3 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.0 | GO:0035729 | cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 1.2 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 2.6 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.3 | GO:0061368 | negative regulation of growth hormone secretion(GO:0060125) behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.6 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 1.7 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 1.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.9 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.2 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 2.3 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 4.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 2.3 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.5 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.3 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 0.6 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 1.4 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.5 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.5 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 1.9 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.9 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 1.0 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 1.9 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.9 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 7.5 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 1.7 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 1.5 | GO:0034968 | histone lysine methylation(GO:0034968) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 1.5 | 13.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.1 | 11.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.7 | 9.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.5 | 7.8 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.4 | 18.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.4 | 3.6 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.4 | 1.5 | GO:0044753 | amphisome(GO:0044753) |
| 0.4 | 3.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.4 | 1.4 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.3 | 6.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 10.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 1.0 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.2 | 4.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.2 | 1.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.2 | 1.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.2 | 18.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.9 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.1 | 18.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 3.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 3.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.7 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 2.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 3.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.3 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 6.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 5.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 4.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 1.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.0 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 2.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.3 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 3.8 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 1.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 2.3 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 7.4 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 4.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 3.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.2 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 6.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 2.1 | 6.3 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) |
| 2.0 | 5.9 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 1.9 | 5.6 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.6 | 11.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 1.5 | 13.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 1.3 | 8.0 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 1.1 | 4.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.9 | 6.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.7 | 4.7 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.6 | 1.9 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.6 | 1.8 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.6 | 17.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.4 | 3.6 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.4 | 2.1 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.4 | 1.7 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.4 | 2.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.4 | 3.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.3 | 1.3 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.3 | 1.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 1.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.3 | 6.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 9.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.3 | 3.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 7.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 1.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 6.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.2 | 1.2 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.2 | 2.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.2 | 1.2 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 3.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 18.2 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.2 | 9.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 0.9 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.2 | 1.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 1.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.6 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.6 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 3.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 7.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 2.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 4.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.0 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.6 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.1 | 10.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 6.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.2 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.1 | 2.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 7.3 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 1.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 2.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 1.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 5.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 0.2 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.1 | 0.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 1.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 1.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 1.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 5.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 1.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 5.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 2.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 1.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 1.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 3.6 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.6 | GO:0071617 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 1.7 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 3.6 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 12.3 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 1.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 2.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 11.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 18.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 6.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 8.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 13.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 7.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 10.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 5.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 3.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 8.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 7.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 4.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.7 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 3.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 2.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 8.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.4 | 5.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 6.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.2 | 10.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 10.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 5.9 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.2 | 13.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 12.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.2 | 6.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 2.5 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 12.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 3.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 2.7 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 4.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 3.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 6.9 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 1.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.4 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 1.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 1.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.8 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 3.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 7.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 3.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 2.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 8.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 3.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 3.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |