Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
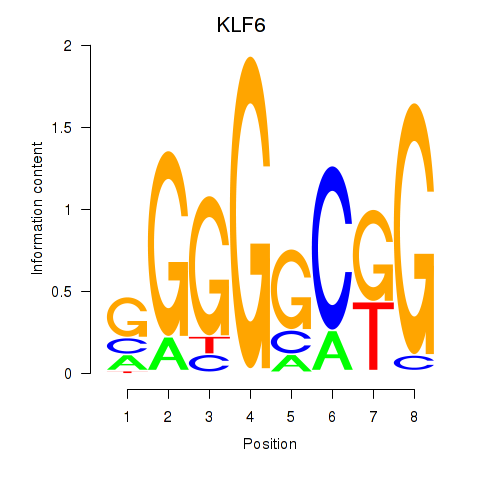
Results for KLF6
Z-value: 1.76
Transcription factors associated with KLF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF6
|
ENSG00000067082.10 | Kruppel like factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF6 | hg19_v2_chr10_-_3827417_3827473 | 0.76 | 9.7e-05 | Click! |
Activity profile of KLF6 motif
Sorted Z-values of KLF6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_57365150 | 7.95 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr1_-_182361327 | 7.13 |
ENST00000331872.6
ENST00000311223.5 |
GLUL
|
glutamate-ammonia ligase |
| chr1_-_6321035 | 7.03 |
ENST00000377893.2
|
GPR153
|
G protein-coupled receptor 153 |
| chr16_+_68679193 | 7.00 |
ENST00000581171.1
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental) |
| chr16_+_68678739 | 6.75 |
ENST00000264012.4
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental) |
| chr1_-_182360498 | 6.54 |
ENST00000417584.2
|
GLUL
|
glutamate-ammonia ligase |
| chr16_+_68678892 | 6.50 |
ENST00000429102.2
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental) |
| chr1_-_182360918 | 6.34 |
ENST00000339526.4
|
GLUL
|
glutamate-ammonia ligase |
| chr7_-_143105941 | 6.26 |
ENST00000275815.3
|
EPHA1
|
EPH receptor A1 |
| chr3_+_50192499 | 5.90 |
ENST00000413852.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr3_+_50192457 | 5.62 |
ENST00000414301.1
ENST00000450338.1 |
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr3_+_50192537 | 5.50 |
ENST00000002829.3
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr5_+_137801160 | 4.22 |
ENST00000239938.4
|
EGR1
|
early growth response 1 |
| chr15_+_98503922 | 4.18 |
ENST00000268042.6
|
ARRDC4
|
arrestin domain containing 4 |
| chr3_+_50192833 | 4.06 |
ENST00000426511.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr21_-_28217721 | 3.85 |
ENST00000284984.3
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr15_+_41136216 | 3.65 |
ENST00000562057.1
ENST00000344051.4 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr14_+_94640633 | 3.41 |
ENST00000304338.3
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4 |
| chr2_+_70142232 | 3.39 |
ENST00000540449.1
|
MXD1
|
MAX dimerization protein 1 |
| chr15_+_41136586 | 3.39 |
ENST00000431806.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr1_+_43148059 | 3.38 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr22_+_31608219 | 3.30 |
ENST00000406516.1
ENST00000444929.2 ENST00000331728.4 |
LIMK2
|
LIM domain kinase 2 |
| chr1_+_43148625 | 3.28 |
ENST00000436427.1
|
YBX1
|
Y box binding protein 1 |
| chr11_+_129939811 | 3.09 |
ENST00000345598.5
ENST00000338167.5 |
APLP2
|
amyloid beta (A4) precursor-like protein 2 |
| chr2_+_70142189 | 3.07 |
ENST00000264444.2
|
MXD1
|
MAX dimerization protein 1 |
| chr11_+_129939779 | 3.05 |
ENST00000533195.1
ENST00000533713.1 ENST00000528499.1 ENST00000539648.1 ENST00000263574.5 |
APLP2
|
amyloid beta (A4) precursor-like protein 2 |
| chr1_+_68150744 | 3.02 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr14_+_94640671 | 2.90 |
ENST00000328839.3
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4 |
| chr11_-_94964354 | 2.87 |
ENST00000536441.1
|
SESN3
|
sestrin 3 |
| chr10_+_72972281 | 2.74 |
ENST00000335350.6
|
UNC5B
|
unc-5 homolog B (C. elegans) |
| chr10_-_25012597 | 2.54 |
ENST00000396432.2
|
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr3_-_113415441 | 2.53 |
ENST00000491165.1
ENST00000316407.4 |
KIAA2018
|
KIAA2018 |
| chr17_+_7341586 | 2.52 |
ENST00000575235.1
|
FGF11
|
fibroblast growth factor 11 |
| chr11_-_65640071 | 2.49 |
ENST00000526624.1
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2 |
| chr11_+_64008525 | 2.48 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr4_-_77819002 | 2.47 |
ENST00000334306.2
|
SOWAHB
|
sosondowah ankyrin repeat domain family member B |
| chr15_+_41136369 | 2.43 |
ENST00000563656.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr1_-_111746966 | 2.41 |
ENST00000369752.5
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr15_-_74726283 | 2.36 |
ENST00000543145.2
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chr15_+_96873921 | 2.35 |
ENST00000394166.3
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr1_-_156051789 | 2.34 |
ENST00000532414.2
|
MEX3A
|
mex-3 RNA binding family member A |
| chr4_-_36246060 | 2.32 |
ENST00000303965.4
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr2_-_220408430 | 2.29 |
ENST00000243776.6
|
CHPF
|
chondroitin polymerizing factor |
| chr10_+_112631699 | 2.28 |
ENST00000444997.1
|
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor) |
| chr11_-_94964210 | 2.24 |
ENST00000416495.2
ENST00000393234.1 |
SESN3
|
sestrin 3 |
| chr6_+_31588478 | 2.19 |
ENST00000376007.4
ENST00000376033.2 |
PRRC2A
|
proline-rich coiled-coil 2A |
| chr12_-_54785074 | 2.18 |
ENST00000338010.5
ENST00000550774.1 |
ZNF385A
|
zinc finger protein 385A |
| chr3_+_184032283 | 2.16 |
ENST00000346169.2
ENST00000414031.1 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_+_202995611 | 2.13 |
ENST00000367240.2
|
PPFIA4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr17_+_79008940 | 2.10 |
ENST00000392411.3
ENST00000575989.1 ENST00000321280.7 ENST00000428708.2 ENST00000575712.1 ENST00000575245.1 ENST00000435091.3 ENST00000321300.6 |
BAIAP2
|
BAI1-associated protein 2 |
| chr6_+_37787704 | 2.08 |
ENST00000474522.1
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr9_+_34989638 | 2.07 |
ENST00000453597.3
ENST00000335998.3 ENST00000312316.5 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr3_+_184032313 | 2.07 |
ENST00000392537.2
ENST00000444134.1 ENST00000450424.1 ENST00000421110.1 ENST00000382330.3 ENST00000426123.1 ENST00000350481.5 ENST00000455679.1 ENST00000440448.1 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr6_+_37137939 | 2.06 |
ENST00000373509.5
|
PIM1
|
pim-1 oncogene |
| chr6_+_44095347 | 2.06 |
ENST00000323267.6
|
TMEM63B
|
transmembrane protein 63B |
| chr19_+_13049413 | 2.04 |
ENST00000316448.5
ENST00000588454.1 |
CALR
|
calreticulin |
| chr5_-_2751762 | 2.04 |
ENST00000302057.5
ENST00000382611.6 |
IRX2
|
iroquois homeobox 2 |
| chr19_-_49568311 | 1.98 |
ENST00000595857.1
ENST00000451356.2 |
NTF4
|
neurotrophin 4 |
| chr7_+_90225796 | 1.94 |
ENST00000380050.3
|
CDK14
|
cyclin-dependent kinase 14 |
| chr3_-_18486354 | 1.91 |
ENST00000493952.2
ENST00000440737.1 |
SATB1
|
SATB homeobox 1 |
| chr4_-_153456153 | 1.90 |
ENST00000603548.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr4_+_89444961 | 1.89 |
ENST00000513325.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr1_-_221915418 | 1.88 |
ENST00000323825.3
ENST00000366899.3 |
DUSP10
|
dual specificity phosphatase 10 |
| chr6_+_43739697 | 1.85 |
ENST00000230480.6
|
VEGFA
|
vascular endothelial growth factor A |
| chr19_+_10196981 | 1.84 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr6_-_35656712 | 1.83 |
ENST00000357266.4
ENST00000542713.1 |
FKBP5
|
FK506 binding protein 5 |
| chr6_-_33285505 | 1.81 |
ENST00000431845.2
|
ZBTB22
|
zinc finger and BTB domain containing 22 |
| chr1_-_113498616 | 1.80 |
ENST00000433570.4
ENST00000538576.1 ENST00000458229.1 |
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr1_-_23670813 | 1.79 |
ENST00000374612.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr22_+_35653445 | 1.75 |
ENST00000420166.1
ENST00000444518.2 ENST00000455359.1 ENST00000216106.5 |
HMGXB4
|
HMG box domain containing 4 |
| chr22_+_41777927 | 1.75 |
ENST00000266304.4
|
TEF
|
thyrotrophic embryonic factor |
| chr1_-_23670752 | 1.74 |
ENST00000302271.6
ENST00000426846.2 ENST00000427764.2 ENST00000606561.1 ENST00000374616.3 |
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr10_-_75634219 | 1.74 |
ENST00000305762.7
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr5_-_81046904 | 1.73 |
ENST00000515395.1
|
SSBP2
|
single-stranded DNA binding protein 2 |
| chr12_-_54785054 | 1.73 |
ENST00000352268.6
ENST00000549962.1 |
ZNF385A
|
zinc finger protein 385A |
| chr19_+_50084561 | 1.71 |
ENST00000246794.5
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr1_+_40506255 | 1.66 |
ENST00000421589.1
|
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr10_-_101380121 | 1.66 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr17_+_45771420 | 1.65 |
ENST00000578982.1
|
TBKBP1
|
TBK1 binding protein 1 |
| chr17_+_8339340 | 1.65 |
ENST00000580012.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr1_-_154531095 | 1.64 |
ENST00000292211.4
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr6_+_33168189 | 1.63 |
ENST00000444757.1
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr1_-_23670817 | 1.59 |
ENST00000478691.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr8_-_57123815 | 1.59 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr6_-_35656685 | 1.56 |
ENST00000539068.1
ENST00000540787.1 |
FKBP5
|
FK506 binding protein 5 |
| chr6_-_110501200 | 1.54 |
ENST00000392586.1
ENST00000419252.1 ENST00000392589.1 ENST00000392588.1 ENST00000359451.2 |
WASF1
|
WAS protein family, member 1 |
| chr11_+_64009072 | 1.53 |
ENST00000535135.1
ENST00000394540.3 |
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr1_+_160175201 | 1.50 |
ENST00000368076.1
|
PEA15
|
phosphoprotein enriched in astrocytes 15 |
| chr10_-_75634326 | 1.49 |
ENST00000322635.3
ENST00000444854.2 ENST00000423381.1 ENST00000322680.3 ENST00000394762.2 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr17_+_47572647 | 1.48 |
ENST00000172229.3
|
NGFR
|
nerve growth factor receptor |
| chr12_+_56473628 | 1.48 |
ENST00000549282.1
ENST00000549061.1 ENST00000267101.3 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr7_+_143078652 | 1.46 |
ENST00000354434.4
ENST00000449423.2 |
ZYX
|
zyxin |
| chr1_+_40506392 | 1.43 |
ENST00000414893.1
ENST00000414281.1 ENST00000420216.1 ENST00000372792.2 ENST00000372798.1 ENST00000340450.3 ENST00000372805.3 ENST00000435719.1 ENST00000427843.1 ENST00000417287.1 ENST00000424977.1 ENST00000446031.1 |
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr4_+_48343339 | 1.42 |
ENST00000264313.6
|
SLAIN2
|
SLAIN motif family, member 2 |
| chr1_+_160175117 | 1.41 |
ENST00000360472.4
|
PEA15
|
phosphoprotein enriched in astrocytes 15 |
| chr21_+_43639211 | 1.41 |
ENST00000450121.1
ENST00000398449.3 ENST00000361802.2 |
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr19_+_7895074 | 1.40 |
ENST00000270530.4
|
EVI5L
|
ecotropic viral integration site 5-like |
| chr10_+_23983671 | 1.38 |
ENST00000376462.1
|
KIAA1217
|
KIAA1217 |
| chr11_-_118661588 | 1.38 |
ENST00000534980.1
ENST00000526070.2 |
DDX6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr1_-_226924980 | 1.37 |
ENST00000272117.3
|
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr17_-_47841485 | 1.36 |
ENST00000506156.1
ENST00000240364.2 |
FAM117A
|
family with sequence similarity 117, member A |
| chr12_-_50222187 | 1.35 |
ENST00000335999.6
|
NCKAP5L
|
NCK-associated protein 5-like |
| chr10_-_75634260 | 1.34 |
ENST00000372765.1
ENST00000351293.3 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr20_+_60813535 | 1.34 |
ENST00000358053.2
ENST00000313733.3 ENST00000439951.2 |
OSBPL2
|
oxysterol binding protein-like 2 |
| chr17_+_48611853 | 1.34 |
ENST00000507709.1
ENST00000515126.1 ENST00000507467.1 |
EPN3
|
epsin 3 |
| chr6_+_31926857 | 1.33 |
ENST00000375394.2
ENST00000544581.1 |
SKIV2L
|
superkiller viralicidic activity 2-like (S. cerevisiae) |
| chr1_-_113498943 | 1.33 |
ENST00000369626.3
|
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr19_-_14629224 | 1.33 |
ENST00000254322.2
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr15_-_70388943 | 1.33 |
ENST00000559048.1
ENST00000560939.1 ENST00000440567.3 ENST00000557907.1 ENST00000558379.1 ENST00000451782.2 ENST00000559929.1 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr1_+_107682629 | 1.31 |
ENST00000370074.4
ENST00000370073.2 ENST00000370071.2 ENST00000542803.1 ENST00000370061.3 ENST00000370072.3 ENST00000370070.2 |
NTNG1
|
netrin G1 |
| chr6_+_83777374 | 1.31 |
ENST00000349129.2
ENST00000237163.5 ENST00000536812.1 |
DOPEY1
|
dopey family member 1 |
| chr15_+_41136263 | 1.31 |
ENST00000568823.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr17_-_42277203 | 1.29 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr20_-_40247002 | 1.29 |
ENST00000373222.3
|
CHD6
|
chromodomain helicase DNA binding protein 6 |
| chr13_-_52027134 | 1.28 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr7_-_129592471 | 1.27 |
ENST00000473814.2
ENST00000490974.1 |
UBE2H
|
ubiquitin-conjugating enzyme E2H |
| chr2_+_220299547 | 1.27 |
ENST00000312358.7
|
SPEG
|
SPEG complex locus |
| chr14_+_21945335 | 1.27 |
ENST00000262709.3
ENST00000457430.2 ENST00000448790.2 |
TOX4
|
TOX high mobility group box family member 4 |
| chr10_-_103874692 | 1.26 |
ENST00000361198.5
|
LDB1
|
LIM domain binding 1 |
| chr1_-_156698181 | 1.26 |
ENST00000313146.6
|
ISG20L2
|
interferon stimulated exonuclease gene 20kDa-like 2 |
| chr12_+_57482665 | 1.25 |
ENST00000300131.3
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr10_+_11060004 | 1.25 |
ENST00000542579.1
ENST00000399850.3 ENST00000417956.2 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr16_+_2563871 | 1.24 |
ENST00000330398.4
ENST00000568562.1 ENST00000569317.1 |
ATP6V0C
ATP6C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c Uncharacterized protein |
| chr2_+_219081817 | 1.24 |
ENST00000315717.5
ENST00000420104.1 ENST00000295685.10 |
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr12_+_14518598 | 1.22 |
ENST00000261168.4
ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr5_-_81046922 | 1.22 |
ENST00000514493.1
ENST00000320672.4 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr11_+_65686952 | 1.21 |
ENST00000527119.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chrX_+_21959108 | 1.21 |
ENST00000457085.1
|
SMS
|
spermine synthase |
| chr6_-_110500905 | 1.21 |
ENST00000392587.2
|
WASF1
|
WAS protein family, member 1 |
| chr11_+_68228186 | 1.20 |
ENST00000393799.2
ENST00000393800.2 ENST00000528635.1 ENST00000533127.1 ENST00000529907.1 ENST00000529344.1 ENST00000534534.1 ENST00000524845.1 ENST00000265637.4 ENST00000524904.1 ENST00000393801.3 ENST00000265636.5 ENST00000529710.1 |
PPP6R3
|
protein phosphatase 6, regulatory subunit 3 |
| chr15_-_43882353 | 1.20 |
ENST00000453080.1
ENST00000360301.4 ENST00000360135.4 ENST00000417085.1 ENST00000431962.1 ENST00000334933.4 ENST00000381879.4 ENST00000420765.1 |
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr6_+_44095263 | 1.20 |
ENST00000532634.1
|
TMEM63B
|
transmembrane protein 63B |
| chr14_+_23791159 | 1.20 |
ENST00000557702.1
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr10_+_45869652 | 1.18 |
ENST00000542434.1
ENST00000374391.2 |
ALOX5
|
arachidonate 5-lipoxygenase |
| chr9_-_34376851 | 1.18 |
ENST00000297625.7
|
KIAA1161
|
KIAA1161 |
| chr1_-_156698591 | 1.16 |
ENST00000368219.1
|
ISG20L2
|
interferon stimulated exonuclease gene 20kDa-like 2 |
| chr3_+_133293278 | 1.16 |
ENST00000508481.1
ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3
|
CDV3 homolog (mouse) |
| chr17_+_42081914 | 1.15 |
ENST00000293404.3
ENST00000589767.1 |
NAGS
|
N-acetylglutamate synthase |
| chr7_+_70229899 | 1.15 |
ENST00000443672.1
|
AUTS2
|
autism susceptibility candidate 2 |
| chr20_+_35201993 | 1.14 |
ENST00000373872.4
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr12_-_49351148 | 1.14 |
ENST00000398092.4
ENST00000539611.1 |
RP11-302B13.5
ARF3
|
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr18_-_51751132 | 1.14 |
ENST00000256429.3
|
MBD2
|
methyl-CpG binding domain protein 2 |
| chr22_+_38004723 | 1.13 |
ENST00000381756.5
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr22_+_38004832 | 1.13 |
ENST00000405147.3
ENST00000429218.1 ENST00000325180.8 ENST00000337437.4 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr4_+_26862313 | 1.13 |
ENST00000467087.1
ENST00000382009.3 ENST00000237364.5 |
STIM2
|
stromal interaction molecule 2 |
| chr1_+_27022839 | 1.12 |
ENST00000457599.2
|
ARID1A
|
AT rich interactive domain 1A (SWI-like) |
| chr4_+_144258021 | 1.12 |
ENST00000262994.4
|
GAB1
|
GRB2-associated binding protein 1 |
| chr19_+_1249869 | 1.12 |
ENST00000591446.2
|
MIDN
|
midnolin |
| chr10_+_11059826 | 1.11 |
ENST00000450189.1
|
CELF2
|
CUGBP, Elav-like family member 2 |
| chr12_-_80328700 | 1.11 |
ENST00000550107.1
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A |
| chr12_+_112451120 | 1.10 |
ENST00000261735.3
ENST00000455836.1 |
ERP29
|
endoplasmic reticulum protein 29 |
| chr1_+_112939121 | 1.10 |
ENST00000441739.1
|
CTTNBP2NL
|
CTTNBP2 N-terminal like |
| chr3_+_49209023 | 1.10 |
ENST00000332780.2
|
KLHDC8B
|
kelch domain containing 8B |
| chr1_+_112938803 | 1.09 |
ENST00000271277.6
|
CTTNBP2NL
|
CTTNBP2 N-terminal like |
| chr8_+_57124245 | 1.09 |
ENST00000521831.1
ENST00000355315.3 ENST00000303759.3 ENST00000517636.1 ENST00000517933.1 ENST00000518801.1 ENST00000523975.1 ENST00000396723.5 ENST00000523061.1 ENST00000521524.1 |
CHCHD7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr3_+_133292759 | 1.07 |
ENST00000431519.2
|
CDV3
|
CDV3 homolog (mouse) |
| chr11_-_65429891 | 1.07 |
ENST00000527874.1
|
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr4_-_103789996 | 1.07 |
ENST00000508238.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr1_-_22263790 | 1.07 |
ENST00000374695.3
|
HSPG2
|
heparan sulfate proteoglycan 2 |
| chr5_+_67511524 | 1.07 |
ENST00000521381.1
ENST00000521657.1 |
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr6_-_150039170 | 1.07 |
ENST00000458696.2
ENST00000392273.3 |
LATS1
|
large tumor suppressor kinase 1 |
| chr6_-_137113604 | 1.06 |
ENST00000359015.4
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr7_+_143079000 | 1.06 |
ENST00000392910.2
|
ZYX
|
zyxin |
| chr7_+_30174668 | 1.06 |
ENST00000415604.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr1_+_6845578 | 1.06 |
ENST00000467404.2
ENST00000439411.2 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr12_-_49351228 | 1.05 |
ENST00000541959.1
ENST00000447318.2 |
ARF3
|
ADP-ribosylation factor 3 |
| chr21_-_40720995 | 1.05 |
ENST00000380749.5
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr5_+_40679584 | 1.04 |
ENST00000302472.3
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr5_-_176900610 | 1.04 |
ENST00000477391.2
ENST00000393565.1 ENST00000309007.5 |
DBN1
|
drebrin 1 |
| chr20_+_35201857 | 1.03 |
ENST00000373874.2
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr6_-_143266297 | 1.03 |
ENST00000367603.2
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr5_+_34929677 | 1.03 |
ENST00000342382.4
ENST00000382021.2 ENST00000303525.7 |
DNAJC21
|
DnaJ (Hsp40) homolog, subfamily C, member 21 |
| chr1_-_234745234 | 1.03 |
ENST00000366610.3
ENST00000366609.3 |
IRF2BP2
|
interferon regulatory factor 2 binding protein 2 |
| chr16_-_30134441 | 1.02 |
ENST00000395200.1
|
MAPK3
|
mitogen-activated protein kinase 3 |
| chr6_+_21593972 | 1.02 |
ENST00000244745.1
ENST00000543472.1 |
SOX4
|
SRY (sex determining region Y)-box 4 |
| chr1_+_33219592 | 1.02 |
ENST00000373481.3
|
KIAA1522
|
KIAA1522 |
| chr18_-_23670546 | 1.01 |
ENST00000542743.1
ENST00000545952.1 ENST00000539849.1 ENST00000415083.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr12_-_31479107 | 1.01 |
ENST00000542983.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr17_+_4736627 | 1.00 |
ENST00000355280.6
ENST00000347992.7 |
MINK1
|
misshapen-like kinase 1 |
| chr11_-_2160180 | 1.00 |
ENST00000381406.4
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr12_+_118814185 | 1.00 |
ENST00000543473.1
|
SUDS3
|
suppressor of defective silencing 3 homolog (S. cerevisiae) |
| chr1_-_91487806 | 0.99 |
ENST00000361321.5
|
ZNF644
|
zinc finger protein 644 |
| chr5_+_60628074 | 0.98 |
ENST00000252744.5
|
ZSWIM6
|
zinc finger, SWIM-type containing 6 |
| chr15_+_44084040 | 0.98 |
ENST00000249786.4
|
SERF2
|
small EDRK-rich factor 2 |
| chr1_+_160175166 | 0.98 |
ENST00000368077.1
|
PEA15
|
phosphoprotein enriched in astrocytes 15 |
| chr14_-_73925225 | 0.98 |
ENST00000356296.4
ENST00000355058.3 ENST00000359560.3 ENST00000557597.1 ENST00000554394.1 ENST00000555238.1 ENST00000535282.1 ENST00000555987.1 ENST00000555394.1 ENST00000554546.1 |
NUMB
|
numb homolog (Drosophila) |
| chr10_+_76586348 | 0.98 |
ENST00000372724.1
ENST00000287239.4 ENST00000372714.1 |
KAT6B
|
K(lysine) acetyltransferase 6B |
| chr16_+_67465016 | 0.98 |
ENST00000326152.5
|
HSD11B2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chrX_+_64887512 | 0.98 |
ENST00000360270.5
|
MSN
|
moesin |
| chr16_-_30102547 | 0.97 |
ENST00000279386.2
|
TBX6
|
T-box 6 |
| chr19_+_532049 | 0.97 |
ENST00000606136.1
|
CDC34
|
cell division cycle 34 |
| chr14_+_23790655 | 0.97 |
ENST00000397276.2
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chrX_+_69674943 | 0.96 |
ENST00000542398.1
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr2_+_159313452 | 0.96 |
ENST00000389757.3
ENST00000389759.3 |
PKP4
|
plakophilin 4 |
| chr11_-_94706705 | 0.96 |
ENST00000279839.6
|
CWC15
|
CWC15 spliceosome-associated protein homolog (S. cerevisiae) |
| chr3_-_47205457 | 0.94 |
ENST00000409792.3
|
SETD2
|
SET domain containing 2 |
| chr1_+_40839369 | 0.94 |
ENST00000372718.3
|
SMAP2
|
small ArfGAP2 |
| chr3_+_184032419 | 0.93 |
ENST00000352767.3
ENST00000427141.2 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_+_117910047 | 0.93 |
ENST00000356554.3
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr1_-_155243235 | 0.93 |
ENST00000355560.4
ENST00000368361.4 |
CLK2
|
CDC-like kinase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 20.2 | GO:1902910 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 5.0 | 20.0 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 1.5 | 21.1 | GO:1901166 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 1.4 | 4.2 | GO:0050720 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) |
| 1.0 | 3.1 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 1.0 | 3.0 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 1.0 | 1.0 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.8 | 2.3 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.7 | 7.9 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.7 | 2.0 | GO:0061193 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.6 | 1.8 | GO:0038190 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) positive regulation of retinal ganglion cell axon guidance(GO:1902336) regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.6 | 1.8 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.6 | 2.4 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.6 | 3.9 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.6 | 1.7 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.6 | 6.7 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.6 | 1.7 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.5 | 2.0 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.5 | 1.4 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.5 | 2.3 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.4 | 1.2 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.4 | 1.9 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.4 | 1.9 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.4 | 1.9 | GO:0030862 | positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.4 | 1.1 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.4 | 1.5 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.4 | 1.1 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.3 | 1.0 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.3 | 5.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.3 | 1.4 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.3 | 2.4 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.3 | 2.7 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.3 | 2.0 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.3 | 1.7 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.3 | 0.9 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.3 | 1.5 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.3 | 5.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.3 | 1.8 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.3 | 1.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.3 | 0.9 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.3 | 0.8 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.3 | 1.0 | GO:0035905 | N-terminal peptidyl-lysine acetylation(GO:0018076) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.3 | 0.8 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.3 | 1.5 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.2 | 0.7 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.2 | 2.7 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.2 | 0.9 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.2 | 2.1 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.2 | 0.7 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.2 | 1.9 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.2 | 3.9 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.2 | 1.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 6.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 10.3 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.2 | 1.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.2 | 3.9 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 0.8 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.2 | 0.8 | GO:1904499 | glial cell fate determination(GO:0007403) negative regulation of mitotic cell cycle, embryonic(GO:0045976) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.2 | 1.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 | 1.3 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.2 | 1.4 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 0.5 | GO:0072034 | primary prostatic bud elongation(GO:0060516) bronchus cartilage development(GO:0060532) common bile duct development(GO:0061009) lung smooth muscle development(GO:0061145) renal vesicle induction(GO:0072034) |
| 0.2 | 1.8 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) progesterone receptor signaling pathway(GO:0050847) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.2 | 1.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.2 | 1.9 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.2 | 1.0 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.2 | 0.5 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 0.5 | GO:0086098 | angiotensin-activated signaling pathway involved in heart process(GO:0086098) negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.2 | 1.7 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.2 | 1.3 | GO:0090166 | Golgi disassembly(GO:0090166) spindle assembly involved in meiosis(GO:0090306) |
| 0.2 | 1.0 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.2 | 5.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 1.3 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.2 | 4.6 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.2 | 2.0 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.2 | 0.5 | GO:0035625 | epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.1 | 5.9 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 0.6 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 | 1.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.4 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.9 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 7.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.5 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 | 0.4 | GO:0045799 | negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 0.7 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 1.2 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.1 | 1.6 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 0.1 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.1 | 0.3 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.1 | 1.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.6 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 0.4 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 1.3 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.1 | 1.4 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 2.4 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 0.5 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 1.0 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 1.4 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 0.9 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 1.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.8 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.5 | GO:0051944 | glial cell-derived neurotrophic factor secretion(GO:0044467) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) negative regulation of dopamine receptor signaling pathway(GO:0060160) regulation of locomotion involved in locomotory behavior(GO:0090325) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.1 | 0.5 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.1 | 0.5 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 1.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 1.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.3 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.1 | 1.0 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.8 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 0.8 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 2.2 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 1.6 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.1 | 0.3 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.8 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.5 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.5 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.1 | 1.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 1.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.9 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 1.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 6.9 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 0.5 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 1.4 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 0.4 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 1.6 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 1.4 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 0.4 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.1 | 0.2 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.1 | 1.0 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) positive regulation of inclusion body assembly(GO:0090261) |
| 0.1 | 0.3 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.1 | 1.0 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.1 | 0.5 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.1 | 0.3 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 4.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.1 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.1 | 1.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 3.1 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 1.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.5 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 1.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.6 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 0.6 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.9 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 1.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.3 | GO:0060028 | somatic muscle development(GO:0007525) non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) convergent extension involved in axis elongation(GO:0060028) axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.3 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) negative regulation of gap junction assembly(GO:1903597) |
| 0.1 | 3.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.4 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 2.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.7 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 1.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.3 | GO:1903299 | regulation of glucokinase activity(GO:0033131) regulation of hexokinase activity(GO:1903299) |
| 0.0 | 0.7 | GO:0072104 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.0 | 0.2 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.2 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.5 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 1.7 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.8 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.2 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 1.0 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.5 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 1.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.4 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 1.2 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 2.1 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 1.8 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.9 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.5 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.2 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.8 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 1.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.3 | GO:0015870 | acetylcholine transport(GO:0015870) |
| 0.0 | 1.6 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.2 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.0 | 1.2 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.7 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.8 | GO:2000146 | negative regulation of locomotion(GO:0040013) negative regulation of cell motility(GO:2000146) |
| 0.0 | 1.5 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 1.6 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.2 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 0.5 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.9 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.7 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.0 | 0.4 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.3 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.5 | GO:0046887 | positive regulation of hormone secretion(GO:0046887) |
| 0.0 | 1.9 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 1.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 2.6 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 2.5 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.4 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.3 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 5.4 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 3.4 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.2 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.2 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.8 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 1.0 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.3 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.4 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.2 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.2 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.5 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.2 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.2 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 1.2 | GO:0051057 | positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.6 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.1 | GO:0030167 | proteoglycan catabolic process(GO:0030167) heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.8 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.6 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) establishment of integrated proviral latency(GO:0075713) |
| 0.0 | 0.2 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 0.4 | GO:0010827 | regulation of glucose transport(GO:0010827) regulation of glucose import(GO:0046324) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.9 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.3 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.4 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.0 | 2.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 2.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.6 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.5 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 1.9 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.0 | 0.2 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 1.1 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 1.3 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.3 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.9 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.3 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.6 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 1.0 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.7 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.1 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 1.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0042482 | positive regulation of odontogenesis(GO:0042482) positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.6 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:0032098 | regulation of appetite(GO:0032098) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 20.9 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.7 | 6.7 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.4 | 1.2 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.4 | 1.1 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.3 | 1.3 | GO:0055087 | Ski complex(GO:0055087) |
| 0.3 | 1.7 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.3 | 1.9 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.3 | 0.9 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.3 | 5.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.3 | 1.9 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.2 | 2.0 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.2 | 1.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 0.8 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.2 | 5.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 3.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.2 | 0.5 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.2 | 1.8 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 1.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 6.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 1.8 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 3.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.8 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 2.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.3 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 0.4 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) gamma-catenin-TCF7L2 complex(GO:0071665) |
| 0.1 | 1.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 1.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 1.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 15.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 1.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 10.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 0.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.4 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.4 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 0.7 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 2.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.5 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.7 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 4.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 1.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 3.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 2.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 3.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 1.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.8 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.8 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 5.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.6 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 1.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 1.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.8 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 1.1 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 2.7 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 2.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.6 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 9.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.2 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 1.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.5 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 2.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.4 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.9 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 2.8 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 4.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.0 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.1 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 1.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 20.0 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 1.0 | 3.1 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.8 | 20.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.8 | 4.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.8 | 2.3 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.6 | 6.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.5 | 1.4 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.4 | 1.7 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.4 | 2.7 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.3 | 1.3 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.3 | 1.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.3 | 1.8 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.3 | 1.1 | GO:0008184 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 0.3 | 1.3 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.2 | 2.4 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.2 | 1.9 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 5.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 1.9 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 1.4 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.2 | 1.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.2 | 2.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 0.7 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.2 | 7.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 1.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 4.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 0.6 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.2 | 0.5 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.2 | 3.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 24.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 1.5 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.2 | 0.5 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.2 | 0.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 0.6 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.2 | 0.6 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 2.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.2 | 0.5 | GO:0051380 | alpha2-adrenergic receptor activity(GO:0004938) norepinephrine binding(GO:0051380) |
| 0.1 | 1.9 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.7 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 1.9 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 1.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.7 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.9 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 1.0 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.9 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.8 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.4 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.1 | 0.7 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 1.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.5 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.1 | 2.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 1.5 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 1.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.0 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 0.4 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.1 | 2.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.5 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 0.4 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.1 | 0.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 5.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 1.6 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 0.3 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 0.9 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 1.2 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.8 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 1.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.4 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.1 | 0.2 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 1.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 1.7 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 1.8 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 2.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 2.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.1 | 0.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.9 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 0.4 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 10.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 0.9 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.7 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.4 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 1.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 2.9 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 1.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 1.6 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 0.5 | GO:0016416 | O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 1.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 6.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.5 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 1.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 6.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.1 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 1.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 1.1 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0003700 | transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 2.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.2 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.2 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 2.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.8 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 3.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0031711 | peptidyl-dipeptidase activity(GO:0008241) bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 1.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 1.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.9 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 1.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 2.7 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 1.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 1.9 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.9 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.8 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 2.6 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 2.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 1.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 1.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.8 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.0 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.9 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.1 | GO:0004104 | acetylcholinesterase activity(GO:0003990) cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 2.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 2.6 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.3 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 2.8 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.9 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.7 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.2 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.4 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 3.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 5.4 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 6.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 8.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 5.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 5.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 21.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 0.9 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 2.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 4.9 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 2.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 9.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 1.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 6.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 2.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 2.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 2.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 8.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 2.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.5 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.3 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 18.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.4 | 7.9 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 18.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 1.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 5.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 2.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 3.6 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.1 | 3.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.6 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 2.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 2.9 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.0 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 0.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 2.9 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 4.4 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 0.5 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.1 | 1.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 2.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 2.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 4.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 9.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 2.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.7 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 1.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 3.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 3.1 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 1.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.9 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 2.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.4 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.5 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |