Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
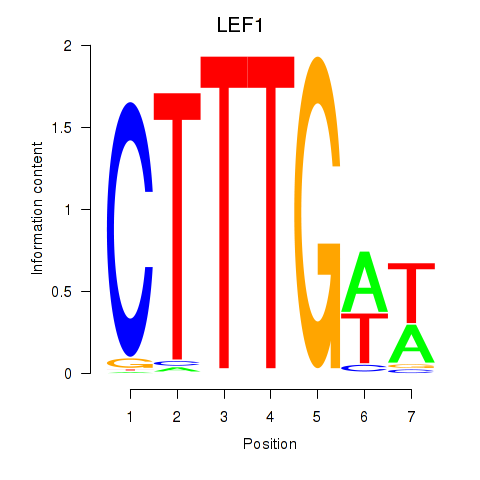
Results for LEF1
Z-value: 1.06
Transcription factors associated with LEF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LEF1
|
ENSG00000138795.5 | lymphoid enhancer binding factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LEF1 | hg19_v2_chr4_-_109087906_109087953 | 0.74 | 1.7e-04 | Click! |
Activity profile of LEF1 motif
Sorted Z-values of LEF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_29027696 | 4.31 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr8_-_124553437 | 4.26 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr16_+_68678892 | 4.25 |
ENST00000429102.2
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental) |
| chr16_+_68679193 | 4.20 |
ENST00000581171.1
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental) |
| chr16_+_68678739 | 3.94 |
ENST00000264012.4
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental) |
| chr2_-_1748214 | 3.41 |
ENST00000433670.1
ENST00000425171.1 ENST00000252804.4 |
PXDN
|
peroxidasin homolog (Drosophila) |
| chr15_+_96869165 | 3.04 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr11_-_94965667 | 2.91 |
ENST00000542176.1
ENST00000278499.2 |
SESN3
|
sestrin 3 |
| chr16_+_55600580 | 2.83 |
ENST00000457326.2
|
CAPNS2
|
calpain, small subunit 2 |
| chr3_+_189349162 | 2.81 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chrX_-_48328631 | 2.79 |
ENST00000429543.1
ENST00000317669.5 |
SLC38A5
|
solute carrier family 38, member 5 |
| chr2_+_14772810 | 2.69 |
ENST00000295092.2
ENST00000331243.4 |
FAM84A
|
family with sequence similarity 84, member A |
| chr7_+_20370300 | 2.46 |
ENST00000537992.1
|
ITGB8
|
integrin, beta 8 |
| chr1_-_26680570 | 2.38 |
ENST00000475866.2
|
AIM1L
|
absent in melanoma 1-like |
| chr2_-_161056762 | 2.36 |
ENST00000428609.2
ENST00000409967.2 |
ITGB6
|
integrin, beta 6 |
| chrX_-_48328551 | 2.36 |
ENST00000376876.3
|
SLC38A5
|
solute carrier family 38, member 5 |
| chr17_-_39093672 | 2.30 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr2_-_161056802 | 2.30 |
ENST00000283249.2
ENST00000409872.1 |
ITGB6
|
integrin, beta 6 |
| chr9_+_71819927 | 2.25 |
ENST00000535702.1
|
TJP2
|
tight junction protein 2 |
| chr5_+_131593364 | 2.19 |
ENST00000253754.3
ENST00000379018.3 |
PDLIM4
|
PDZ and LIM domain 4 |
| chr15_+_41136216 | 2.13 |
ENST00000562057.1
ENST00000344051.4 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr17_+_75447326 | 2.09 |
ENST00000591088.1
|
SEPT9
|
septin 9 |
| chr16_-_4987065 | 2.03 |
ENST00000590782.2
ENST00000345988.2 |
PPL
|
periplakin |
| chr15_+_41136734 | 1.99 |
ENST00000568580.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr11_-_87908600 | 1.99 |
ENST00000531138.1
ENST00000526372.1 ENST00000243662.6 |
RAB38
|
RAB38, member RAS oncogene family |
| chr9_+_71820057 | 1.98 |
ENST00000539225.1
|
TJP2
|
tight junction protein 2 |
| chr14_-_23624511 | 1.97 |
ENST00000529705.2
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr7_-_41742697 | 1.91 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr5_+_31193847 | 1.87 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr15_+_43885252 | 1.86 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr11_-_125351481 | 1.86 |
ENST00000577924.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr1_+_78383813 | 1.85 |
ENST00000342754.5
|
NEXN
|
nexilin (F actin binding protein) |
| chr3_+_107244229 | 1.85 |
ENST00000456419.1
ENST00000402163.2 |
BBX
|
bobby sox homolog (Drosophila) |
| chr18_+_20494078 | 1.83 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr3_+_152017924 | 1.80 |
ENST00000465907.2
ENST00000492948.1 ENST00000485509.1 ENST00000464596.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr15_+_43985084 | 1.78 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr8_+_102504979 | 1.77 |
ENST00000395927.1
|
GRHL2
|
grainyhead-like 2 (Drosophila) |
| chrX_-_32173579 | 1.72 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr15_+_43985725 | 1.64 |
ENST00000413453.2
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr21_+_39628852 | 1.58 |
ENST00000398938.2
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chrX_-_80457385 | 1.56 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr15_-_23932437 | 1.56 |
ENST00000331837.4
|
NDN
|
necdin, melanoma antigen (MAGE) family member |
| chr13_+_76334498 | 1.55 |
ENST00000534657.1
|
LMO7
|
LIM domain 7 |
| chr1_-_116383738 | 1.54 |
ENST00000320238.3
|
NHLH2
|
nescient helix loop helix 2 |
| chr22_-_31688381 | 1.54 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr4_+_146403912 | 1.52 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr1_+_27668505 | 1.50 |
ENST00000318074.5
|
SYTL1
|
synaptotagmin-like 1 |
| chr2_-_9143786 | 1.47 |
ENST00000462696.1
ENST00000305997.3 |
MBOAT2
|
membrane bound O-acyltransferase domain containing 2 |
| chr3_+_182983090 | 1.46 |
ENST00000465010.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr1_+_68150744 | 1.45 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr15_+_41136369 | 1.45 |
ENST00000563656.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chrX_+_56259316 | 1.45 |
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8 |
| chr21_-_42879909 | 1.44 |
ENST00000458356.1
ENST00000398585.3 ENST00000424093.1 |
TMPRSS2
|
transmembrane protease, serine 2 |
| chr7_+_73245193 | 1.44 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr19_+_11071652 | 1.43 |
ENST00000344626.4
ENST00000429416.3 |
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr3_-_18466026 | 1.42 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr3_-_169381183 | 1.38 |
ENST00000494292.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr6_-_31846744 | 1.37 |
ENST00000414427.1
ENST00000229729.6 ENST00000375562.4 |
SLC44A4
|
solute carrier family 44, member 4 |
| chr4_+_30721968 | 1.37 |
ENST00000361762.2
|
PCDH7
|
protocadherin 7 |
| chr1_-_115292591 | 1.36 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr7_-_80548493 | 1.35 |
ENST00000536800.1
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr10_+_24498060 | 1.34 |
ENST00000376454.3
ENST00000376452.3 |
KIAA1217
|
KIAA1217 |
| chr7_+_20370746 | 1.34 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr4_-_140477353 | 1.34 |
ENST00000406354.1
ENST00000506866.2 |
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr15_+_43885799 | 1.33 |
ENST00000449946.1
ENST00000417289.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr1_+_100111580 | 1.31 |
ENST00000605497.1
|
PALMD
|
palmdelphin |
| chr5_+_112074029 | 1.30 |
ENST00000512211.2
|
APC
|
adenomatous polyposis coli |
| chr21_+_39628655 | 1.30 |
ENST00000398925.1
ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr14_+_85996507 | 1.29 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr10_+_11206925 | 1.28 |
ENST00000354440.2
ENST00000315874.4 ENST00000427450.1 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr19_+_11071546 | 1.27 |
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chrX_-_15619076 | 1.26 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr3_-_71353892 | 1.26 |
ENST00000484350.1
|
FOXP1
|
forkhead box P1 |
| chr16_+_57673348 | 1.24 |
ENST00000567915.1
ENST00000564103.1 ENST00000562467.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr3_-_167813132 | 1.22 |
ENST00000309027.4
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr14_+_24837226 | 1.22 |
ENST00000554050.1
ENST00000554903.1 ENST00000554779.1 ENST00000250373.4 ENST00000553708.1 |
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
| chr3_-_141747439 | 1.21 |
ENST00000467667.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr17_-_56065484 | 1.21 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr1_+_47799542 | 1.20 |
ENST00000471289.2
ENST00000450808.2 |
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr18_+_55888767 | 1.20 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr12_-_31477072 | 1.19 |
ENST00000454658.2
|
FAM60A
|
family with sequence similarity 60, member A |
| chr15_+_93443419 | 1.19 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr4_+_78079450 | 1.18 |
ENST00000395640.1
ENST00000512918.1 |
CCNG2
|
cyclin G2 |
| chr3_-_71179699 | 1.18 |
ENST00000497355.1
|
FOXP1
|
forkhead box P1 |
| chr14_+_85996471 | 1.17 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr13_+_38923959 | 1.17 |
ENST00000379649.1
ENST00000239878.4 ENST00000437952.1 ENST00000379641.1 |
UFM1
|
ubiquitin-fold modifier 1 |
| chr1_+_100111479 | 1.16 |
ENST00000263174.4
|
PALMD
|
palmdelphin |
| chrX_+_48367338 | 1.16 |
ENST00000359882.4
ENST00000537758.1 ENST00000367574.4 ENST00000355961.4 ENST00000489940.1 ENST00000361988.3 |
PORCN
|
porcupine homolog (Drosophila) |
| chr15_+_96873921 | 1.15 |
ENST00000394166.3
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr4_+_78078304 | 1.15 |
ENST00000316355.5
ENST00000354403.5 ENST00000502280.1 |
CCNG2
|
cyclin G2 |
| chr10_-_126138622 | 1.15 |
ENST00000451024.3
|
NKX1-2
|
NK1 homeobox 2 |
| chr2_+_27665232 | 1.15 |
ENST00000543753.1
ENST00000288873.3 |
KRTCAP3
|
keratinocyte associated protein 3 |
| chr3_-_112360116 | 1.14 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chr1_+_39571026 | 1.14 |
ENST00000524432.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr4_-_74088800 | 1.13 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr4_+_154387480 | 1.11 |
ENST00000409663.3
ENST00000440693.1 ENST00000409959.3 |
KIAA0922
|
KIAA0922 |
| chr22_-_31688431 | 1.11 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr4_+_78079570 | 1.10 |
ENST00000509972.1
|
CCNG2
|
cyclin G2 |
| chr10_-_115933942 | 1.10 |
ENST00000369285.3
ENST00000369287.3 ENST00000369286.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr4_-_140477910 | 1.09 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr15_+_52043758 | 1.09 |
ENST00000249700.4
ENST00000539962.2 |
TMOD2
|
tropomodulin 2 (neuronal) |
| chr5_+_145317356 | 1.07 |
ENST00000511217.1
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr11_-_57194948 | 1.06 |
ENST00000533235.1
ENST00000526621.1 ENST00000352187.1 |
SLC43A3
|
solute carrier family 43, member 3 |
| chr4_+_113152881 | 1.06 |
ENST00000274000.5
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr3_-_69402828 | 1.05 |
ENST00000460709.1
|
FRMD4B
|
FERM domain containing 4B |
| chr11_-_111794446 | 1.05 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr4_-_157892167 | 1.05 |
ENST00000541126.1
|
PDGFC
|
platelet derived growth factor C |
| chr10_+_24497704 | 1.05 |
ENST00000376456.4
ENST00000458595.1 |
KIAA1217
|
KIAA1217 |
| chr9_+_504674 | 1.04 |
ENST00000382297.2
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr5_+_137673200 | 1.04 |
ENST00000434981.2
|
FAM53C
|
family with sequence similarity 53, member C |
| chr1_+_82266053 | 1.02 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr3_-_66024213 | 1.02 |
ENST00000483466.1
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr1_+_230203010 | 1.02 |
ENST00000541865.1
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr3_-_69591727 | 1.02 |
ENST00000459638.1
|
FRMD4B
|
FERM domain containing 4B |
| chr9_+_112852477 | 1.01 |
ENST00000480388.1
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr2_-_216257849 | 1.01 |
ENST00000456923.1
|
FN1
|
fibronectin 1 |
| chr17_+_29815013 | 1.01 |
ENST00000394744.2
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II) |
| chr3_-_141747950 | 1.01 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr3_-_99569821 | 1.01 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr7_+_90338712 | 1.01 |
ENST00000265741.3
ENST00000406263.1 |
CDK14
|
cyclin-dependent kinase 14 |
| chr8_-_139926236 | 1.00 |
ENST00000303045.6
ENST00000435777.1 |
COL22A1
|
collagen, type XXII, alpha 1 |
| chr11_-_10830463 | 0.99 |
ENST00000527419.1
ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr5_+_138210919 | 0.99 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr2_+_54684327 | 0.98 |
ENST00000389980.3
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr7_-_80548667 | 0.98 |
ENST00000265361.3
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr14_+_52164820 | 0.98 |
ENST00000554167.1
|
FRMD6
|
FERM domain containing 6 |
| chr3_+_177159695 | 0.96 |
ENST00000442937.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr11_+_101983176 | 0.95 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr14_-_21270995 | 0.94 |
ENST00000555698.1
ENST00000397970.4 ENST00000340900.3 |
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr6_-_11779403 | 0.93 |
ENST00000414691.3
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr13_-_39612176 | 0.93 |
ENST00000352251.3
ENST00000350125.3 |
PROSER1
|
proline and serine rich 1 |
| chr15_+_52043813 | 0.93 |
ENST00000435126.2
|
TMOD2
|
tropomodulin 2 (neuronal) |
| chr1_-_9953295 | 0.93 |
ENST00000377258.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr3_-_71179988 | 0.93 |
ENST00000491238.1
|
FOXP1
|
forkhead box P1 |
| chr10_-_95241951 | 0.93 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr11_-_8892464 | 0.92 |
ENST00000527347.1
ENST00000526241.1 ENST00000526126.1 ENST00000530938.1 ENST00000526057.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr11_+_110001723 | 0.92 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr2_+_201171268 | 0.92 |
ENST00000423749.1
ENST00000428692.1 ENST00000457757.1 ENST00000453663.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr14_+_52164675 | 0.91 |
ENST00000555936.1
|
FRMD6
|
FERM domain containing 6 |
| chr10_+_114710425 | 0.91 |
ENST00000352065.5
ENST00000369395.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr2_+_27665289 | 0.91 |
ENST00000407293.1
|
KRTCAP3
|
keratinocyte associated protein 3 |
| chr6_+_32939964 | 0.91 |
ENST00000607833.1
|
BRD2
|
bromodomain containing 2 |
| chr8_-_17104099 | 0.90 |
ENST00000524358.1
|
CNOT7
|
CCR4-NOT transcription complex, subunit 7 |
| chr12_+_1738363 | 0.90 |
ENST00000397196.2
|
WNT5B
|
wingless-type MMTV integration site family, member 5B |
| chr16_+_57673207 | 0.90 |
ENST00000564783.1
ENST00000564729.1 ENST00000565976.1 ENST00000566508.1 ENST00000544297.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr4_+_76649753 | 0.90 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr10_-_30024716 | 0.90 |
ENST00000375398.2
ENST00000375400.3 |
SVIL
|
supervillin |
| chr2_+_66666432 | 0.90 |
ENST00000495021.2
|
MEIS1
|
Meis homeobox 1 |
| chr4_+_76649797 | 0.90 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr4_-_157892498 | 0.89 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C |
| chr11_-_3013482 | 0.89 |
ENST00000529361.1
ENST00000528968.1 ENST00000534372.1 ENST00000531291.1 ENST00000526842.1 |
NAP1L4
|
nucleosome assembly protein 1-like 4 |
| chr8_+_42873548 | 0.89 |
ENST00000533338.1
ENST00000534420.1 |
HOOK3
RP11-598P20.5
|
hook microtubule-tethering protein 3 Uncharacterized protein |
| chr1_+_93913713 | 0.88 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr15_+_93447675 | 0.88 |
ENST00000536619.1
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr3_+_182983126 | 0.87 |
ENST00000481531.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr10_-_116164239 | 0.87 |
ENST00000419268.1
ENST00000304129.4 ENST00000545353.1 |
AFAP1L2
|
actin filament associated protein 1-like 2 |
| chr14_-_67981916 | 0.87 |
ENST00000357461.2
|
TMEM229B
|
transmembrane protein 229B |
| chr1_-_93645818 | 0.86 |
ENST00000370280.1
ENST00000479918.1 |
TMED5
|
transmembrane emp24 protein transport domain containing 5 |
| chr5_+_3596168 | 0.86 |
ENST00000302006.3
|
IRX1
|
iroquois homeobox 1 |
| chr5_+_67576062 | 0.86 |
ENST00000523807.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr11_-_57194817 | 0.86 |
ENST00000529748.1
ENST00000525474.1 |
SLC43A3
|
solute carrier family 43, member 3 |
| chr4_-_157892055 | 0.86 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr17_-_46035187 | 0.86 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr18_-_53089723 | 0.85 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr13_+_98086445 | 0.85 |
ENST00000245304.4
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr5_+_66124590 | 0.85 |
ENST00000490016.2
ENST00000403666.1 ENST00000450827.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr8_-_128231299 | 0.84 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr8_-_42358742 | 0.84 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr11_+_69455855 | 0.84 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr1_+_12538594 | 0.84 |
ENST00000543710.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr10_-_95242044 | 0.84 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr15_+_52311398 | 0.83 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr12_-_76817036 | 0.83 |
ENST00000546946.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr21_+_37692481 | 0.83 |
ENST00000400485.1
|
MORC3
|
MORC family CW-type zinc finger 3 |
| chr1_+_59762642 | 0.83 |
ENST00000371218.4
ENST00000303721.7 |
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr12_+_104359641 | 0.83 |
ENST00000537100.1
|
TDG
|
thymine-DNA glycosylase |
| chr1_+_174844645 | 0.83 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr17_+_75449889 | 0.83 |
ENST00000590938.1
|
SEPT9
|
septin 9 |
| chr10_+_11047259 | 0.83 |
ENST00000379261.4
ENST00000416382.2 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr4_-_146859787 | 0.82 |
ENST00000508784.1
|
ZNF827
|
zinc finger protein 827 |
| chr3_+_111578583 | 0.82 |
ENST00000478922.1
ENST00000477695.1 |
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr14_-_105444694 | 0.82 |
ENST00000333244.5
|
AHNAK2
|
AHNAK nucleoprotein 2 |
| chr13_-_99630233 | 0.82 |
ENST00000376460.1
ENST00000442173.1 |
DOCK9
|
dedicator of cytokinesis 9 |
| chrX_+_44732757 | 0.82 |
ENST00000377967.4
ENST00000536777.1 ENST00000382899.4 ENST00000543216.1 |
KDM6A
|
lysine (K)-specific demethylase 6A |
| chr13_+_76210448 | 0.81 |
ENST00000377499.5
|
LMO7
|
LIM domain 7 |
| chr4_+_26165074 | 0.80 |
ENST00000512351.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr4_+_160203650 | 0.80 |
ENST00000514565.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr1_-_161337662 | 0.80 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr1_-_23670752 | 0.80 |
ENST00000302271.6
ENST00000426846.2 ENST00000427764.2 ENST00000606561.1 ENST00000374616.3 |
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr11_+_67007518 | 0.80 |
ENST00000530342.1
ENST00000308783.5 |
KDM2A
|
lysine (K)-specific demethylase 2A |
| chr21_-_32931290 | 0.80 |
ENST00000286827.3
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1 |
| chr3_+_141105235 | 0.80 |
ENST00000503809.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr19_+_13134772 | 0.80 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr1_-_115053781 | 0.80 |
ENST00000358465.2
ENST00000369543.2 |
TRIM33
|
tripartite motif containing 33 |
| chr6_+_4776580 | 0.79 |
ENST00000397588.3
|
CDYL
|
chromodomain protein, Y-like |
| chr1_-_156722195 | 0.79 |
ENST00000368206.5
|
HDGF
|
hepatoma-derived growth factor |
| chr1_+_16085244 | 0.79 |
ENST00000400773.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr11_-_3013316 | 0.79 |
ENST00000430811.1
|
NAP1L4
|
nucleosome assembly protein 1-like 4 |
| chr7_-_27169801 | 0.79 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr14_-_38064198 | 0.79 |
ENST00000250448.2
|
FOXA1
|
forkhead box A1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of LEF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.4 | GO:0060901 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 1.0 | 4.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.7 | 2.8 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.6 | 1.9 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.6 | 3.7 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.6 | 1.7 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.5 | 2.7 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.5 | 2.6 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.5 | 5.7 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.5 | 1.8 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.5 | 1.8 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.4 | 4.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.4 | 2.5 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.4 | 1.2 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.4 | 1.1 | GO:0070632 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.3 | 1.0 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.3 | 1.0 | GO:1903450 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.3 | 8.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.3 | 2.4 | GO:0048625 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.3 | 0.6 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.3 | 5.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.3 | 2.0 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.3 | 1.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.3 | 0.8 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.3 | 0.8 | GO:0007388 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.3 | 1.3 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.2 | 1.7 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 1.7 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 2.0 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 0.7 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.2 | 4.3 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.2 | 1.1 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.2 | 1.3 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.2 | 0.9 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.2 | 2.3 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.2 | 1.9 | GO:0003383 | apical constriction(GO:0003383) |
| 0.2 | 3.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 1.0 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 2.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 1.7 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.2 | 1.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.2 | 0.6 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.2 | 0.9 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.2 | 0.7 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.2 | 2.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 2.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 1.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 | 0.5 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.2 | 1.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 | 2.9 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 0.5 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 1.2 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 0.5 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.2 | 5.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.6 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.7 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 0.4 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 1.2 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 | 0.8 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.1 | 0.4 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 1.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.4 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.1 | 1.4 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 1.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.7 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.8 | GO:1903786 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.1 | 0.8 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 0.8 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.4 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.1 | 0.7 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.9 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 1.1 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.1 | 2.8 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.3 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.3 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.1 | 0.6 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.3 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.1 | 1.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.6 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.4 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 | 0.5 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 0.5 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.5 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 0.5 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.1 | 0.3 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 0.8 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.6 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.1 | 0.5 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 3.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 1.5 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 1.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.7 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.3 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 4.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.8 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 1.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.3 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.9 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.7 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.4 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 0.2 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.1 | 0.4 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 1.0 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 3.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 1.7 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.1 | 0.8 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 0.4 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 0.7 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 0.9 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 | 0.9 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 1.8 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 0.4 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 2.1 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.1 | 0.3 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 | 0.3 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.4 | GO:0000436 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.1 | 0.8 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 1.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.3 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 0.9 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 3.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 2.0 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 0.9 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 1.5 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 0.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 1.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.3 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 1.8 | GO:0060674 | placenta blood vessel development(GO:0060674) |
| 0.1 | 2.1 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 1.8 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.3 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 0.4 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.8 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.8 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.8 | GO:0033327 | Leydig cell differentiation(GO:0033327) response to UV-A(GO:0070141) |
| 0.1 | 0.6 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.2 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.3 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.7 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 1.9 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 2.7 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 1.9 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.4 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.6 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.7 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 1.9 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.5 | GO:0003264 | cardioblast proliferation(GO:0003263) regulation of cardioblast proliferation(GO:0003264) |
| 0.0 | 1.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.5 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.6 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 1.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.8 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.2 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.4 | GO:0090181 | regulation of cholesterol biosynthetic process(GO:0045540) regulation of cholesterol metabolic process(GO:0090181) |
| 0.0 | 0.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.8 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.3 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 | 0.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.5 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:2000843 | testosterone secretion(GO:0035936) regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.5 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 5.2 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 1.6 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.7 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 1.1 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:1904814 | regulation of protein localization to chromosome, telomeric region(GO:1904814) positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.0 | 1.3 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.6 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 2.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.3 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.3 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 | 0.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.5 | GO:0060711 | labyrinthine layer development(GO:0060711) |
| 0.0 | 0.1 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 0.1 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.7 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0033522 | histone H2A ubiquitination(GO:0033522) histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.6 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 1.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.4 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 1.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 1.3 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.0 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 2.7 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 2.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 1.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.5 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.8 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.2 | GO:0070779 | gamma-aminobutyric acid biosynthetic process(GO:0009449) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.3 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.2 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 2.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 1.6 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.2 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.1 | GO:0021831 | embryonic olfactory bulb interneuron precursor migration(GO:0021831) chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.0 | 0.4 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 1.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.7 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.0 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.5 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) |
| 0.0 | 0.2 | GO:0060453 | regulation of gastric acid secretion(GO:0060453) |
| 0.0 | 0.6 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.2 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.2 | GO:2000810 | regulation of bicellular tight junction assembly(GO:2000810) |
| 0.0 | 0.1 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.9 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 3.8 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 1.6 | 4.7 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.6 | 1.9 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.4 | 2.0 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.4 | 1.1 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.4 | 1.1 | GO:0075341 | host cell PML body(GO:0075341) |
| 0.3 | 2.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.3 | 2.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.3 | 0.8 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 1.0 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.2 | 1.7 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.2 | 1.7 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.2 | 2.9 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 0.5 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 6.0 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 6.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 3.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.6 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.1 | 1.0 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.3 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.9 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.6 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.7 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 0.5 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 1.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.6 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 1.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.3 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.2 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.1 | 1.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.7 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 11.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 3.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 1.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.7 | GO:0032059 | bleb(GO:0032059) |
| 0.1 | 1.7 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 0.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.6 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 1.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 1.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.6 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 1.2 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 2.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 9.2 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 2.0 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 1.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 1.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 3.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.0 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 1.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.6 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 1.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 1.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 1.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.9 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 1.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.8 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.7 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.7 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 2.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 2.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.6 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.5 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 4.8 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 3.4 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 1.5 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 3.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.9 | 3.7 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.9 | 2.6 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.7 | 3.4 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.6 | 1.8 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.5 | 1.5 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.5 | 0.5 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.5 | 2.3 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.4 | 1.8 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.4 | 1.2 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.3 | 5.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.3 | 3.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.3 | 1.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.2 | 1.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 1.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 0.8 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.2 | 2.8 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 1.4 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.2 | 1.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.2 | 3.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 2.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.9 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 2.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 3.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.5 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 2.8 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 0.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 1.7 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.7 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 2.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.4 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 0.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.9 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 1.7 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 2.0 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.8 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 0.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.5 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.1 | 0.5 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 1.9 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.8 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 1.8 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.7 | GO:0042835 | BRE binding(GO:0042835) |
| 0.1 | 1.7 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 4.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.6 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.1 | 0.3 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 2.0 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 2.8 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.3 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.3 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.9 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.6 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.1 | 0.3 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.5 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.3 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.5 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 2.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 3.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 2.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.2 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.1 | 1.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 2.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.4 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 0.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 3.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 4.2 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 2.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 1.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 1.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 3.7 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 1.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 2.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.6 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.3 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 4.7 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.6 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 1.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 5.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.4 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 1.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.7 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 1.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.9 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 1.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 1.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 3.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.6 | GO:0008266 | poly-pyrimidine tract binding(GO:0008187) poly(U) RNA binding(GO:0008266) |
| 0.0 | 5.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 1.2 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 1.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 2.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.9 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.0 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 11.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.6 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.0 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.6 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 10.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.0 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 1.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 3.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.6 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.9 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.3 | GO:0047485 | protein N-terminus binding(GO:0047485) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 3.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 1.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 3.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 5.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 6.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 1.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 5.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 4.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 3.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 2.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 2.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 3.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 3.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 2.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.5 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.9 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 12.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 1.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.8 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 4.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 3.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 6.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.5 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 7.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 2.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 2.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.4 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 2.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 3.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 5.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |