Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
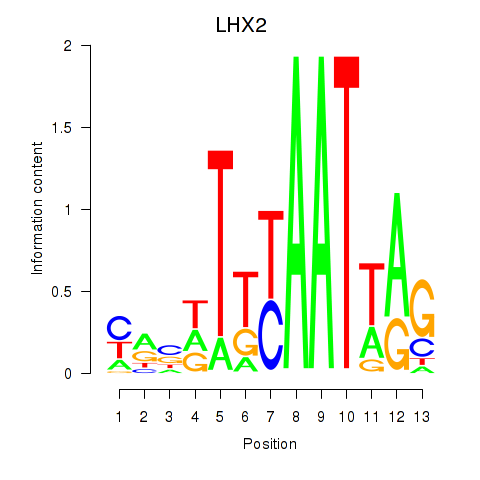
Results for LHX2
Z-value: 0.29
Transcription factors associated with LHX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX2
|
ENSG00000106689.6 | LIM homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX2 | hg19_v2_chr9_+_126777676_126777700 | -0.29 | 2.1e-01 | Click! |
Activity profile of LHX2 motif
Sorted Z-values of LHX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_29027696 | 0.61 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr19_+_42212526 | 0.37 |
ENST00000598976.1
ENST00000435837.2 ENST00000221992.6 ENST00000405816.1 |
CEA
CEACAM5
|
Uncharacterized protein carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr5_-_150473127 | 0.28 |
ENST00000521001.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr19_+_42212501 | 0.24 |
ENST00000398599.4
|
CEACAM5
|
carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr3_-_98619999 | 0.23 |
ENST00000449482.1
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2 |
| chr3_+_57882024 | 0.23 |
ENST00000494088.1
|
SLMAP
|
sarcolemma associated protein |
| chr12_+_41136144 | 0.23 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr3_+_57882061 | 0.22 |
ENST00000461354.1
ENST00000466255.1 |
SLMAP
|
sarcolemma associated protein |
| chr3_+_57881966 | 0.21 |
ENST00000495364.1
|
SLMAP
|
sarcolemma associated protein |
| chr1_+_160709055 | 0.21 |
ENST00000368043.3
ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7
|
SLAM family member 7 |
| chr4_+_71384300 | 0.21 |
ENST00000504451.1
|
AMTN
|
amelotin |
| chr21_-_27423339 | 0.21 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr6_-_138833630 | 0.20 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr15_-_42565023 | 0.20 |
ENST00000566474.1
|
TMEM87A
|
transmembrane protein 87A |
| chr16_+_23652773 | 0.19 |
ENST00000563998.1
ENST00000568589.1 ENST00000568272.1 |
DCTN5
|
dynactin 5 (p25) |
| chr17_-_39216344 | 0.19 |
ENST00000391418.2
|
KRTAP2-3
|
keratin associated protein 2-3 |
| chr4_+_69313145 | 0.19 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr5_+_118668846 | 0.18 |
ENST00000513374.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr12_-_49245916 | 0.18 |
ENST00000552512.1
ENST00000551468.1 |
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr6_-_131291572 | 0.17 |
ENST00000529208.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr6_-_26199499 | 0.17 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr9_-_100854838 | 0.17 |
ENST00000538344.1
|
TRIM14
|
tripartite motif containing 14 |
| chr1_+_41448820 | 0.16 |
ENST00000372616.1
|
CTPS1
|
CTP synthase 1 |
| chr11_-_62323702 | 0.16 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr4_-_90756769 | 0.16 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr17_+_61086917 | 0.15 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr12_+_6309517 | 0.15 |
ENST00000382519.4
ENST00000009180.4 |
CD9
|
CD9 molecule |
| chr6_+_30848771 | 0.15 |
ENST00000503180.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr14_+_56078695 | 0.14 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr1_+_160370344 | 0.14 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr2_+_102953608 | 0.14 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr10_-_29923893 | 0.14 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr3_+_190281229 | 0.13 |
ENST00000453359.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr1_+_160709076 | 0.13 |
ENST00000359331.4
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr6_+_30848557 | 0.13 |
ENST00000460944.2
ENST00000324771.8 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr1_-_183560011 | 0.12 |
ENST00000367536.1
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr1_+_160709029 | 0.12 |
ENST00000444090.2
ENST00000441662.2 |
SLAMF7
|
SLAM family member 7 |
| chr1_+_207238326 | 0.12 |
ENST00000541914.1
|
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr15_-_72521017 | 0.12 |
ENST00000561609.1
|
PKM
|
pyruvate kinase, muscle |
| chr6_-_26199471 | 0.12 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr9_+_108320392 | 0.12 |
ENST00000602661.1
ENST00000223528.2 ENST00000448551.2 ENST00000540160.1 |
FKTN
|
fukutin |
| chr17_+_45000483 | 0.12 |
ENST00000576910.2
ENST00000439730.2 ENST00000393456.2 ENST00000415811.2 ENST00000575949.1 ENST00000225567.4 ENST00000572403.1 ENST00000570879.1 |
GOSR2
|
golgi SNAP receptor complex member 2 |
| chr4_-_153332886 | 0.11 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr11_-_46113756 | 0.11 |
ENST00000531959.1
|
PHF21A
|
PHD finger protein 21A |
| chr14_+_100532771 | 0.11 |
ENST00000557153.1
|
EVL
|
Enah/Vasp-like |
| chr1_+_84630574 | 0.11 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr17_-_38859996 | 0.11 |
ENST00000264651.2
|
KRT24
|
keratin 24 |
| chr1_+_152975488 | 0.11 |
ENST00000542696.1
|
SPRR3
|
small proline-rich protein 3 |
| chrX_+_105936982 | 0.11 |
ENST00000418562.1
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr1_+_87012922 | 0.11 |
ENST00000263723.5
|
CLCA4
|
chloride channel accessory 4 |
| chr21_+_35014706 | 0.11 |
ENST00000399353.1
ENST00000444491.1 ENST00000381318.3 |
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr16_+_23652700 | 0.10 |
ENST00000300087.2
|
DCTN5
|
dynactin 5 (p25) |
| chr18_+_34124507 | 0.10 |
ENST00000591635.1
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr3_+_38017264 | 0.10 |
ENST00000436654.1
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr14_-_51027838 | 0.10 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr14_+_67708344 | 0.10 |
ENST00000557237.1
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr2_-_37501692 | 0.09 |
ENST00000443977.1
|
PRKD3
|
protein kinase D3 |
| chr1_-_232651312 | 0.09 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr14_-_68159152 | 0.09 |
ENST00000554035.1
|
RDH11
|
retinol dehydrogenase 11 (all-trans/9-cis/11-cis) |
| chr8_+_92082424 | 0.09 |
ENST00000285420.4
ENST00000404789.3 |
OTUD6B
|
OTU domain containing 6B |
| chr21_+_35014783 | 0.08 |
ENST00000381291.4
ENST00000381285.4 ENST00000399367.3 ENST00000399352.1 ENST00000399355.2 ENST00000399349.1 |
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr5_+_137722255 | 0.08 |
ENST00000542866.1
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr21_+_34619079 | 0.08 |
ENST00000433395.2
|
AP000295.9
|
AP000295.9 |
| chr7_+_101460882 | 0.08 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr16_+_88636875 | 0.08 |
ENST00000569435.1
|
ZC3H18
|
zinc finger CCCH-type containing 18 |
| chr11_-_104972158 | 0.07 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr2_+_29001711 | 0.07 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr1_-_168464875 | 0.07 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr21_-_43735628 | 0.07 |
ENST00000291525.10
ENST00000518498.1 |
TFF3
|
trefoil factor 3 (intestinal) |
| chr9_+_116207007 | 0.07 |
ENST00000374140.2
|
RGS3
|
regulator of G-protein signaling 3 |
| chr16_+_31213206 | 0.07 |
ENST00000561916.2
|
C16orf98
|
chromosome 16 open reading frame 98 |
| chr14_-_78083112 | 0.07 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr5_-_41794313 | 0.07 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr11_-_108093329 | 0.07 |
ENST00000278612.8
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus |
| chr10_-_116286656 | 0.06 |
ENST00000428430.1
ENST00000369266.3 ENST00000392952.3 |
ABLIM1
|
actin binding LIM protein 1 |
| chr16_-_18887627 | 0.06 |
ENST00000563235.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr3_-_131221771 | 0.06 |
ENST00000510154.1
ENST00000507669.1 |
MRPL3
|
mitochondrial ribosomal protein L3 |
| chr4_-_57253587 | 0.06 |
ENST00000513376.1
ENST00000602986.1 ENST00000434343.2 ENST00000451613.1 ENST00000205214.6 ENST00000502617.1 |
AASDH
|
aminoadipate-semialdehyde dehydrogenase |
| chr14_-_23395623 | 0.06 |
ENST00000556043.1
|
PRMT5
|
protein arginine methyltransferase 5 |
| chrX_+_105937068 | 0.06 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr1_+_52682052 | 0.06 |
ENST00000371591.1
|
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr6_+_130339710 | 0.06 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr2_+_234627424 | 0.06 |
ENST00000373409.3
|
UGT1A4
|
UDP glucuronosyltransferase 1 family, polypeptide A4 |
| chr1_-_85870177 | 0.06 |
ENST00000542148.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr11_+_27076764 | 0.05 |
ENST00000525090.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr4_-_89951028 | 0.05 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr20_+_56964169 | 0.05 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr13_+_24844819 | 0.05 |
ENST00000399949.2
|
SPATA13
|
spermatogenesis associated 13 |
| chr10_-_61900762 | 0.05 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr19_+_20188830 | 0.05 |
ENST00000418063.2
|
ZNF90
|
zinc finger protein 90 |
| chr2_+_210517895 | 0.05 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr9_-_125240235 | 0.05 |
ENST00000259357.2
|
OR1J1
|
olfactory receptor, family 1, subfamily J, member 1 |
| chr5_+_98109322 | 0.05 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr2_-_112237835 | 0.05 |
ENST00000442293.1
ENST00000439494.1 |
MIR4435-1HG
|
MIR4435-1 host gene (non-protein coding) |
| chr2_+_145780725 | 0.05 |
ENST00000451478.1
|
TEX41
|
testis expressed 41 (non-protein coding) |
| chr5_-_78281775 | 0.04 |
ENST00000396151.3
ENST00000565165.1 |
ARSB
|
arylsulfatase B |
| chr1_+_62439037 | 0.04 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr12_-_118796971 | 0.04 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr9_-_77643307 | 0.04 |
ENST00000376834.3
ENST00000376830.3 |
C9orf41
|
chromosome 9 open reading frame 41 |
| chr20_+_57464200 | 0.04 |
ENST00000604005.1
|
GNAS
|
GNAS complex locus |
| chr13_+_24844857 | 0.04 |
ENST00000409126.1
ENST00000343003.6 |
SPATA13
|
spermatogenesis associated 13 |
| chrY_+_15418467 | 0.04 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr17_+_17082842 | 0.04 |
ENST00000579361.1
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr16_-_23652570 | 0.04 |
ENST00000261584.4
|
PALB2
|
partner and localizer of BRCA2 |
| chr3_-_160823040 | 0.04 |
ENST00000484127.1
ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr2_-_175712479 | 0.04 |
ENST00000443238.1
|
CHN1
|
chimerin 1 |
| chr12_+_6554021 | 0.04 |
ENST00000266557.3
|
CD27
|
CD27 molecule |
| chr8_+_28748765 | 0.04 |
ENST00000355231.5
|
HMBOX1
|
homeobox containing 1 |
| chr2_-_128284020 | 0.04 |
ENST00000295321.4
ENST00000455721.2 |
IWS1
|
IWS1 homolog (S. cerevisiae) |
| chr17_-_62499334 | 0.04 |
ENST00000579996.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr3_+_49977894 | 0.03 |
ENST00000433811.1
|
RBM6
|
RNA binding motif protein 6 |
| chr2_-_231084617 | 0.03 |
ENST00000409815.2
|
SP110
|
SP110 nuclear body protein |
| chr7_-_151217166 | 0.03 |
ENST00000496004.1
|
RHEB
|
Ras homolog enriched in brain |
| chr13_+_24844979 | 0.03 |
ENST00000454083.1
|
SPATA13
|
spermatogenesis associated 13 |
| chr12_+_128399965 | 0.03 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr19_-_44384291 | 0.03 |
ENST00000324394.6
|
ZNF404
|
zinc finger protein 404 |
| chr12_-_118796910 | 0.03 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr2_-_166702601 | 0.03 |
ENST00000428888.1
|
AC009495.4
|
AC009495.4 |
| chr3_-_12587055 | 0.03 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chrX_-_20074895 | 0.03 |
ENST00000543767.1
|
MAP7D2
|
MAP7 domain containing 2 |
| chr16_-_66864806 | 0.03 |
ENST00000566336.1
ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr11_-_26744908 | 0.03 |
ENST00000533617.1
|
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr21_+_35014829 | 0.03 |
ENST00000451686.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr1_+_145883868 | 0.03 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chr2_+_87135076 | 0.03 |
ENST00000409776.2
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr5_-_159546396 | 0.02 |
ENST00000523662.1
ENST00000456329.3 ENST00000307063.7 |
PWWP2A
|
PWWP domain containing 2A |
| chr3_-_160823158 | 0.02 |
ENST00000392779.2
ENST00000392780.1 ENST00000494173.1 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr7_-_103848405 | 0.02 |
ENST00000447452.2
ENST00000545943.1 ENST00000297431.4 |
ORC5
|
origin recognition complex, subunit 5 |
| chr4_-_185694872 | 0.02 |
ENST00000505492.1
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr4_-_109541539 | 0.02 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr3_-_145940214 | 0.02 |
ENST00000481701.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr8_-_37411648 | 0.02 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr16_+_4606347 | 0.02 |
ENST00000444310.4
|
C16orf96
|
chromosome 16 open reading frame 96 |
| chr5_-_156536126 | 0.02 |
ENST00000522593.1
|
HAVCR2
|
hepatitis A virus cellular receptor 2 |
| chr4_+_147096837 | 0.02 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr11_-_40315640 | 0.02 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr12_+_128399917 | 0.02 |
ENST00000544645.1
|
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr13_+_73632897 | 0.02 |
ENST00000377687.4
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr19_-_30199516 | 0.02 |
ENST00000591243.1
|
C19orf12
|
chromosome 19 open reading frame 12 |
| chr2_+_68962014 | 0.02 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr13_+_33160553 | 0.02 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr3_-_131221790 | 0.02 |
ENST00000512877.1
ENST00000264995.3 ENST00000511168.1 ENST00000425847.2 |
MRPL3
|
mitochondrial ribosomal protein L3 |
| chr17_-_34308524 | 0.02 |
ENST00000293275.3
|
CCL16
|
chemokine (C-C motif) ligand 16 |
| chr17_-_39140549 | 0.02 |
ENST00000377755.4
|
KRT40
|
keratin 40 |
| chr2_+_145780767 | 0.02 |
ENST00000599358.1
ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr3_-_100551141 | 0.01 |
ENST00000478235.1
ENST00000471901.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr12_-_118797475 | 0.01 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr20_+_30697298 | 0.01 |
ENST00000398022.2
|
TM9SF4
|
transmembrane 9 superfamily protein member 4 |
| chr11_+_31391381 | 0.01 |
ENST00000465995.1
ENST00000536040.1 |
DNAJC24
|
DnaJ (Hsp40) homolog, subfamily C, member 24 |
| chr21_-_43735446 | 0.01 |
ENST00000398431.2
|
TFF3
|
trefoil factor 3 (intestinal) |
| chr10_-_14372870 | 0.01 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chr4_-_1670632 | 0.01 |
ENST00000461064.1
|
FAM53A
|
family with sequence similarity 53, member A |
| chr6_-_66417107 | 0.01 |
ENST00000370621.3
ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS
|
eyes shut homolog (Drosophila) |
| chr3_-_129513259 | 0.01 |
ENST00000329333.5
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr14_-_106926724 | 0.01 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr5_+_68860949 | 0.01 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr7_-_99716940 | 0.01 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr8_-_109260897 | 0.01 |
ENST00000521297.1
ENST00000519030.1 ENST00000521440.1 ENST00000518345.1 ENST00000519627.1 ENST00000220849.5 |
EIF3E
|
eukaryotic translation initiation factor 3, subunit E |
| chr1_-_147142358 | 0.01 |
ENST00000392988.2
|
ACP6
|
acid phosphatase 6, lysophosphatidic |
| chr9_-_98268883 | 0.01 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chr2_+_68961905 | 0.01 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr10_+_96443204 | 0.01 |
ENST00000339022.5
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr6_-_76072719 | 0.01 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chrM_+_12331 | 0.01 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr3_-_107099454 | 0.01 |
ENST00000593837.1
ENST00000599431.1 |
RP11-446H18.5
|
RP11-446H18.5 |
| chr6_+_26199737 | 0.01 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chr17_-_57229155 | 0.01 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr2_-_55496174 | 0.01 |
ENST00000417363.1
ENST00000412530.1 ENST00000394600.3 ENST00000366137.2 ENST00000420637.1 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr7_-_99716914 | 0.01 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr2_+_68961934 | 0.01 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr7_-_65113280 | 0.01 |
ENST00000593865.1
|
AC104057.1
|
Protein LOC100996407 |
| chr3_+_62936098 | 0.00 |
ENST00000475886.1
ENST00000465684.1 ENST00000465262.1 ENST00000468072.1 |
LINC00698
|
long intergenic non-protein coding RNA 698 |
| chr2_-_89157161 | 0.00 |
ENST00000390237.2
|
IGKC
|
immunoglobulin kappa constant |
| chr12_+_122688090 | 0.00 |
ENST00000324189.4
ENST00000546192.1 |
B3GNT4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr7_-_122840015 | 0.00 |
ENST00000194130.2
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporter), member 1 |
| chr3_+_119187785 | 0.00 |
ENST00000295588.4
ENST00000476573.1 |
POGLUT1
|
protein O-glucosyltransferase 1 |
| chr7_+_80253387 | 0.00 |
ENST00000438020.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr1_+_210501589 | 0.00 |
ENST00000413764.2
ENST00000541565.1 |
HHAT
|
hedgehog acyltransferase |
| chr16_+_333152 | 0.00 |
ENST00000219406.6
ENST00000404312.1 ENST00000456379.1 |
PDIA2
|
protein disulfide isomerase family A, member 2 |
| chr1_+_244515930 | 0.00 |
ENST00000366537.1
ENST00000308105.4 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr13_-_51101468 | 0.00 |
ENST00000428276.1
|
RP11-175B12.2
|
RP11-175B12.2 |
| chr6_+_126661253 | 0.00 |
ENST00000368326.1
ENST00000368325.1 ENST00000368328.4 |
CENPW
|
centromere protein W |
| chr4_-_186570679 | 0.00 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_+_145780662 | 0.00 |
ENST00000423031.1
|
TEX41
|
testis expressed 41 (non-protein coding) |
Network of associatons between targets according to the STRING database.
First level regulatory network of LHX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.2 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.1 | 0.3 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.1 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.2 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.0 | 0.2 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.3 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.0 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.2 | GO:1903284 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.6 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.2 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.2 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.0 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.2 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.6 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) protease inhibitor complex(GO:0097179) |
| 0.0 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |