Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
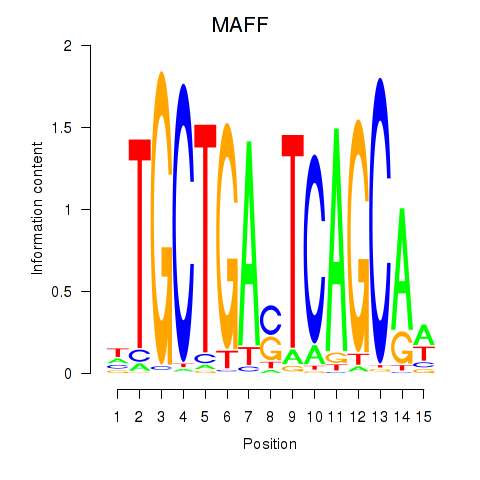
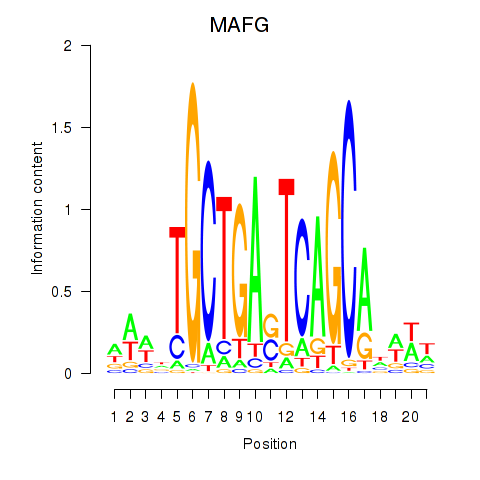
Results for MAFF_MAFG
Z-value: 0.83
Transcription factors associated with MAFF_MAFG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAFF
|
ENSG00000185022.7 | MAF bZIP transcription factor F |
|
MAFG
|
ENSG00000197063.6 | MAF bZIP transcription factor G |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MAFF | hg19_v2_chr22_+_38609538_38609547 | -0.80 | 1.9e-05 | Click! |
| MAFG | hg19_v2_chr17_-_79885576_79885624 | 0.67 | 1.2e-03 | Click! |
Activity profile of MAFF_MAFG motif
Sorted Z-values of MAFF_MAFG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrY_+_2709527 | 10.07 |
ENST00000250784.8
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1 |
| chrY_+_2709906 | 3.83 |
ENST00000430575.1
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1 |
| chr12_+_20968608 | 3.45 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr18_-_70931689 | 3.01 |
ENST00000581862.1
|
RP11-169F17.1
|
Protein LOC400655 |
| chr8_-_95229531 | 2.58 |
ENST00000450165.2
|
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr11_-_117695449 | 2.26 |
ENST00000292079.2
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr7_-_86849883 | 2.24 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr11_+_62652649 | 2.21 |
ENST00000539507.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr2_-_86094764 | 2.02 |
ENST00000393808.3
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr16_+_66442411 | 1.86 |
ENST00000499966.1
|
LINC00920
|
long intergenic non-protein coding RNA 920 |
| chr11_-_114271139 | 1.85 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chr17_+_18625336 | 1.72 |
ENST00000395671.4
ENST00000571542.1 ENST00000395672.2 ENST00000414850.2 ENST00000424146.2 |
TRIM16L
|
tripartite motif containing 16-like |
| chr7_-_86849836 | 1.66 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr2_+_74781828 | 1.65 |
ENST00000340004.6
|
DOK1
|
docking protein 1, 62kDa (downstream of tyrosine kinase 1) |
| chr12_+_56075330 | 1.64 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr1_-_238108575 | 1.62 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr17_-_79881408 | 1.57 |
ENST00000392366.3
|
MAFG
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr3_-_150264272 | 1.56 |
ENST00000491660.1
ENST00000487153.1 ENST00000239944.2 |
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr1_-_24438664 | 1.51 |
ENST00000374434.3
ENST00000330966.7 ENST00000329601.7 |
MYOM3
|
myomesin 3 |
| chrX_-_14891150 | 1.46 |
ENST00000452869.1
ENST00000398334.1 ENST00000324138.3 |
FANCB
|
Fanconi anemia, complementation group B |
| chr9_+_114423615 | 1.44 |
ENST00000374293.4
|
GNG10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr10_+_47083454 | 1.41 |
ENST00000374312.1
|
NPY4R
|
neuropeptide Y receptor Y4 |
| chr12_+_47610315 | 1.37 |
ENST00000548348.1
ENST00000549500.1 |
PCED1B
|
PC-esterase domain containing 1B |
| chr1_+_17248418 | 1.36 |
ENST00000375541.5
|
CROCC
|
ciliary rootlet coiled-coil, rootletin |
| chr19_-_33555780 | 1.32 |
ENST00000254260.3
ENST00000400226.4 |
RHPN2
|
rhophilin, Rho GTPase binding protein 2 |
| chr20_+_61287711 | 1.32 |
ENST00000370507.1
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1 |
| chr19_+_39421556 | 1.26 |
ENST00000407800.2
ENST00000402029.3 |
MRPS12
|
mitochondrial ribosomal protein S12 |
| chr10_+_5005445 | 1.25 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr2_+_46844290 | 1.23 |
ENST00000238892.3
|
CRIPT
|
cysteine-rich PDZ-binding protein |
| chr2_+_120189422 | 1.22 |
ENST00000306406.4
|
TMEM37
|
transmembrane protein 37 |
| chr4_-_170924888 | 1.20 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr5_+_139027877 | 1.16 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chr6_+_27215494 | 1.16 |
ENST00000230582.3
|
PRSS16
|
protease, serine, 16 (thymus) |
| chr4_-_185655278 | 1.16 |
ENST00000281453.5
|
MLF1IP
|
centromere protein U |
| chr1_+_240408560 | 1.15 |
ENST00000441342.1
ENST00000545751.1 |
FMN2
|
formin 2 |
| chr17_-_63822563 | 1.15 |
ENST00000317442.8
|
CEP112
|
centrosomal protein 112kDa |
| chr8_-_27469383 | 1.14 |
ENST00000519742.1
|
CLU
|
clusterin |
| chr20_+_61867235 | 1.13 |
ENST00000342412.6
ENST00000217169.3 |
BIRC7
|
baculoviral IAP repeat containing 7 |
| chr12_+_111051902 | 1.12 |
ENST00000397655.3
ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1
|
tectonic family member 1 |
| chr3_-_142720267 | 1.12 |
ENST00000597953.1
|
RP11-91G21.1
|
RP11-91G21.1 |
| chr6_+_159291090 | 1.11 |
ENST00000367073.4
ENST00000608817.1 |
C6orf99
|
chromosome 6 open reading frame 99 |
| chr9_-_14693417 | 1.09 |
ENST00000380916.4
|
ZDHHC21
|
zinc finger, DHHC-type containing 21 |
| chr7_-_150777920 | 1.07 |
ENST00000353841.2
ENST00000297532.6 |
FASTK
|
Fas-activated serine/threonine kinase |
| chr11_+_44748361 | 1.06 |
ENST00000533202.1
ENST00000533080.1 ENST00000520358.2 ENST00000520999.2 |
TSPAN18
|
tetraspanin 18 |
| chr19_-_49955050 | 1.05 |
ENST00000262265.5
|
PIH1D1
|
PIH1 domain containing 1 |
| chr17_+_9728828 | 1.05 |
ENST00000262441.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr7_-_150777874 | 1.02 |
ENST00000540185.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr11_-_60010556 | 1.00 |
ENST00000427611.2
|
MS4A4E
|
membrane-spanning 4-domains, subfamily A, member 4E |
| chr21_+_27543175 | 1.00 |
ENST00000608591.1
ENST00000609365.1 |
AP000230.1
|
AP000230.1 |
| chr15_+_34517194 | 0.99 |
ENST00000267750.4
ENST00000249209.4 ENST00000561372.1 ENST00000559078.1 ENST00000557879.1 |
EMC4
|
ER membrane protein complex subunit 4 |
| chr6_+_10556215 | 0.98 |
ENST00000316170.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr17_-_34890037 | 0.95 |
ENST00000589404.1
|
MYO19
|
myosin XIX |
| chr2_-_234763147 | 0.94 |
ENST00000411486.2
ENST00000432087.1 ENST00000441687.1 ENST00000414924.1 |
HJURP
|
Holliday junction recognition protein |
| chr19_-_39421377 | 0.94 |
ENST00000430193.3
ENST00000600042.1 ENST00000221431.6 |
SARS2
|
seryl-tRNA synthetase 2, mitochondrial |
| chr19_+_48876300 | 0.93 |
ENST00000600863.1
ENST00000601610.1 ENST00000595322.1 |
SYNGR4
|
synaptogyrin 4 |
| chr1_+_174968564 | 0.92 |
ENST00000426793.1
|
CACYBP
|
calcyclin binding protein |
| chr12_+_48592134 | 0.92 |
ENST00000595310.1
|
DKFZP779L1853
|
DKFZP779L1853 |
| chr10_+_54074033 | 0.92 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr4_+_3768075 | 0.91 |
ENST00000509482.1
ENST00000330055.5 |
ADRA2C
|
adrenoceptor alpha 2C |
| chrX_-_10544942 | 0.91 |
ENST00000380779.1
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr10_-_14590644 | 0.90 |
ENST00000378470.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr7_-_150777949 | 0.90 |
ENST00000482571.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr16_+_30211181 | 0.90 |
ENST00000395138.2
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr17_+_60758814 | 0.90 |
ENST00000579432.1
ENST00000446119.2 |
MRC2
|
mannose receptor, C type 2 |
| chr1_+_10459111 | 0.89 |
ENST00000541529.1
ENST00000270776.8 ENST00000483936.1 ENST00000538557.1 |
PGD
|
phosphogluconate dehydrogenase |
| chr8_-_27850180 | 0.89 |
ENST00000380385.2
ENST00000301906.4 ENST00000354914.3 |
SCARA5
|
scavenger receptor class A, member 5 (putative) |
| chr8_-_95220775 | 0.89 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr5_+_140019079 | 0.87 |
ENST00000252100.6
|
TMCO6
|
transmembrane and coiled-coil domains 6 |
| chr19_-_55791431 | 0.86 |
ENST00000593263.1
ENST00000376343.3 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr1_-_68962744 | 0.86 |
ENST00000525124.1
|
DEPDC1
|
DEP domain containing 1 |
| chr11_+_76839283 | 0.85 |
ENST00000409709.3
ENST00000409893.1 ENST00000458637.2 ENST00000409619.2 |
MYO7A
|
myosin VIIA |
| chr5_-_90610200 | 0.82 |
ENST00000511918.1
ENST00000513626.1 ENST00000607854.1 |
LUCAT1
RP11-213H15.4
|
lung cancer associated transcript 1 (non-protein coding) RP11-213H15.4 |
| chr7_+_139025875 | 0.81 |
ENST00000297534.6
|
C7orf55
|
chromosome 7 open reading frame 55 |
| chr1_-_27998689 | 0.80 |
ENST00000339145.4
ENST00000362020.4 ENST00000361157.6 |
IFI6
|
interferon, alpha-inducible protein 6 |
| chr15_-_66858298 | 0.80 |
ENST00000537670.1
|
LCTL
|
lactase-like |
| chr17_-_14140166 | 0.80 |
ENST00000420162.2
ENST00000431716.2 |
CDRT15
|
CMT1A duplicated region transcript 15 |
| chr10_+_46994087 | 0.80 |
ENST00000374317.1
|
GPRIN2
|
G protein regulated inducer of neurite outgrowth 2 |
| chr16_-_1429627 | 0.78 |
ENST00000248104.7
|
UNKL
|
unkempt family zinc finger-like |
| chr1_+_203830703 | 0.77 |
ENST00000414487.2
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E |
| chr1_+_22351977 | 0.77 |
ENST00000420503.1
ENST00000416769.1 ENST00000404210.2 |
LINC00339
|
long intergenic non-protein coding RNA 339 |
| chr15_-_31521567 | 0.77 |
ENST00000560812.1
ENST00000559853.1 ENST00000558109.1 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr11_-_34938039 | 0.76 |
ENST00000395787.3
|
APIP
|
APAF1 interacting protein |
| chr11_-_85376121 | 0.71 |
ENST00000527447.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr4_-_89080003 | 0.71 |
ENST00000237612.3
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr10_-_134145321 | 0.71 |
ENST00000368625.4
ENST00000368619.3 ENST00000456004.1 ENST00000368620.2 |
STK32C
|
serine/threonine kinase 32C |
| chr7_+_5465382 | 0.70 |
ENST00000609130.1
|
RP11-1275H24.2
|
RP11-1275H24.2 |
| chr11_+_67250490 | 0.70 |
ENST00000528641.2
ENST00000279146.3 |
AIP
|
aryl hydrocarbon receptor interacting protein |
| chr19_+_15751689 | 0.69 |
ENST00000586182.2
ENST00000591058.1 ENST00000221307.8 |
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3 |
| chr15_+_23810903 | 0.68 |
ENST00000564592.1
|
MKRN3
|
makorin ring finger protein 3 |
| chr11_+_72983246 | 0.68 |
ENST00000393590.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr7_-_14026123 | 0.68 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr14_-_58894332 | 0.68 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr10_+_26727125 | 0.67 |
ENST00000376236.4
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr13_+_24844857 | 0.67 |
ENST00000409126.1
ENST00000343003.6 |
SPATA13
|
spermatogenesis associated 13 |
| chr15_-_72668185 | 0.67 |
ENST00000457859.2
ENST00000566304.1 ENST00000567159.1 ENST00000429918.2 |
HEXA
|
hexosaminidase A (alpha polypeptide) |
| chr17_-_76921459 | 0.65 |
ENST00000262768.7
|
TIMP2
|
TIMP metallopeptidase inhibitor 2 |
| chr10_+_88854926 | 0.65 |
ENST00000298784.1
ENST00000298786.4 |
FAM35A
|
family with sequence similarity 35, member A |
| chr15_-_54025300 | 0.65 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr19_-_14168391 | 0.64 |
ENST00000589048.1
|
PALM3
|
paralemmin 3 |
| chr20_-_3748363 | 0.63 |
ENST00000217195.8
|
C20orf27
|
chromosome 20 open reading frame 27 |
| chrX_+_155110956 | 0.63 |
ENST00000286448.6
ENST00000262640.6 ENST00000460621.1 |
VAMP7
|
vesicle-associated membrane protein 7 |
| chr6_+_27215471 | 0.62 |
ENST00000421826.2
|
PRSS16
|
protease, serine, 16 (thymus) |
| chr6_-_127840453 | 0.62 |
ENST00000556132.1
|
SOGA3
|
SOGA family member 3 |
| chr14_-_58893832 | 0.61 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr14_-_58893876 | 0.61 |
ENST00000555097.1
ENST00000555404.1 |
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr14_-_58894223 | 0.60 |
ENST00000555593.1
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr2_-_46844159 | 0.60 |
ENST00000474980.1
ENST00000306465.4 |
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr10_+_43916052 | 0.60 |
ENST00000442526.2
|
RP11-517P14.2
|
RP11-517P14.2 |
| chrX_+_72783026 | 0.59 |
ENST00000373504.6
ENST00000373502.5 |
CHIC1
|
cysteine-rich hydrophobic domain 1 |
| chr16_-_66907139 | 0.59 |
ENST00000561579.2
|
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr19_-_49955096 | 0.59 |
ENST00000595550.1
|
PIH1D1
|
PIH1 domain containing 1 |
| chr4_-_123843597 | 0.58 |
ENST00000510735.1
ENST00000304430.5 |
NUDT6
|
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
| chr17_-_3794021 | 0.57 |
ENST00000381769.2
|
CAMKK1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha |
| chr2_+_74648848 | 0.57 |
ENST00000409791.1
ENST00000426787.1 ENST00000348227.4 |
WDR54
|
WD repeat domain 54 |
| chrX_-_55187588 | 0.57 |
ENST00000472571.2
ENST00000332132.4 ENST00000425133.2 ENST00000358460.4 |
FAM104B
|
family with sequence similarity 104, member B |
| chr3_-_118864893 | 0.57 |
ENST00000354673.2
ENST00000425327.2 |
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr19_-_51014588 | 0.57 |
ENST00000598418.1
|
JOSD2
|
Josephin domain containing 2 |
| chr7_+_89975979 | 0.56 |
ENST00000257659.8
ENST00000222511.6 ENST00000417207.1 |
GTPBP10
|
GTP-binding protein 10 (putative) |
| chr2_-_111435610 | 0.56 |
ENST00000447014.1
ENST00000420328.1 ENST00000535254.1 ENST00000409311.1 ENST00000302759.6 |
BUB1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr19_-_51014460 | 0.56 |
ENST00000595669.1
|
JOSD2
|
Josephin domain containing 2 |
| chr3_-_178969403 | 0.56 |
ENST00000314235.5
ENST00000392685.2 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr5_+_140529630 | 0.56 |
ENST00000543635.1
|
PCDHB6
|
protocadherin beta 6 |
| chr2_+_87808725 | 0.56 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr21_-_47352477 | 0.55 |
ENST00000593412.1
|
PRED62
|
Uncharacterized protein |
| chr6_-_127840336 | 0.54 |
ENST00000525778.1
|
SOGA3
|
SOGA family member 3 |
| chr15_+_49913201 | 0.54 |
ENST00000329873.5
ENST00000558653.1 ENST00000559164.1 ENST00000560632.1 ENST00000559405.1 ENST00000251250.6 |
DTWD1
|
DTW domain containing 1 |
| chr15_+_33010175 | 0.53 |
ENST00000300177.4
ENST00000560677.1 ENST00000560830.1 |
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr15_+_23810853 | 0.53 |
ENST00000568252.1
|
MKRN3
|
makorin ring finger protein 3 |
| chr11_-_10879593 | 0.53 |
ENST00000528289.1
ENST00000432999.2 |
ZBED5
|
zinc finger, BED-type containing 5 |
| chr1_+_161719552 | 0.53 |
ENST00000367943.4
|
DUSP12
|
dual specificity phosphatase 12 |
| chr16_+_78133536 | 0.53 |
ENST00000402655.2
ENST00000406884.2 ENST00000539474.2 ENST00000569818.1 ENST00000355860.3 ENST00000408984.3 |
WWOX
|
WW domain containing oxidoreductase |
| chr2_+_211421262 | 0.52 |
ENST00000233072.5
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr17_-_17485731 | 0.52 |
ENST00000395783.1
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chrX_+_140096761 | 0.51 |
ENST00000370530.1
|
SPANXB1
|
SPANX family, member B1 |
| chr8_+_125486939 | 0.51 |
ENST00000303545.3
|
RNF139
|
ring finger protein 139 |
| chr4_-_101111615 | 0.51 |
ENST00000273990.2
|
DDIT4L
|
DNA-damage-inducible transcript 4-like |
| chr19_-_36297348 | 0.51 |
ENST00000589835.1
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr7_+_100199800 | 0.50 |
ENST00000223061.5
|
PCOLCE
|
procollagen C-endopeptidase enhancer |
| chr2_-_46844242 | 0.49 |
ENST00000281382.6
|
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr6_+_2988199 | 0.49 |
ENST00000450238.1
ENST00000445000.1 ENST00000426637.1 |
LINC01011
NQO2
|
long intergenic non-protein coding RNA 1011 NAD(P)H dehydrogenase, quinone 2 |
| chr10_-_73497581 | 0.49 |
ENST00000398786.2
|
C10orf105
|
chromosome 10 open reading frame 105 |
| chr13_+_37574678 | 0.49 |
ENST00000389704.3
|
EXOSC8
|
exosome component 8 |
| chr11_-_10879572 | 0.49 |
ENST00000413761.2
|
ZBED5
|
zinc finger, BED-type containing 5 |
| chr2_+_172864490 | 0.49 |
ENST00000315796.4
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr2_+_128458514 | 0.48 |
ENST00000310981.4
|
SFT2D3
|
SFT2 domain containing 3 |
| chr14_-_75530693 | 0.48 |
ENST00000555135.1
ENST00000357971.3 ENST00000553302.1 ENST00000555694.1 ENST00000238618.3 |
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr9_-_77502636 | 0.47 |
ENST00000449912.2
|
TRPM6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr17_-_73266616 | 0.47 |
ENST00000579194.1
ENST00000581777.1 |
MIF4GD
|
MIF4G domain containing |
| chr2_-_167232484 | 0.46 |
ENST00000375387.4
ENST00000303354.6 ENST00000409672.1 |
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit |
| chr8_-_98290087 | 0.46 |
ENST00000322128.3
|
TSPYL5
|
TSPY-like 5 |
| chr11_+_57308979 | 0.46 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr5_+_140019004 | 0.46 |
ENST00000394671.3
ENST00000511410.1 ENST00000537378.1 |
TMCO6
|
transmembrane and coiled-coil domains 6 |
| chr12_+_27863706 | 0.46 |
ENST00000081029.3
ENST00000538315.1 ENST00000542791.1 |
MRPS35
|
mitochondrial ribosomal protein S35 |
| chr10_+_124030819 | 0.46 |
ENST00000260723.4
ENST00000368994.2 |
BTBD16
|
BTB (POZ) domain containing 16 |
| chr16_+_31128978 | 0.45 |
ENST00000448516.2
ENST00000219797.4 |
KAT8
|
K(lysine) acetyltransferase 8 |
| chr4_+_156775910 | 0.45 |
ENST00000506072.1
ENST00000507590.1 |
TDO2
|
tryptophan 2,3-dioxygenase |
| chr12_+_101988774 | 0.45 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr1_+_47901689 | 0.45 |
ENST00000334793.5
|
FOXD2
|
forkhead box D2 |
| chr12_-_91573132 | 0.45 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr17_-_28619059 | 0.44 |
ENST00000580709.1
|
BLMH
|
bleomycin hydrolase |
| chr4_+_105828537 | 0.44 |
ENST00000515649.1
|
RP11-556I14.1
|
RP11-556I14.1 |
| chr12_+_121163602 | 0.44 |
ENST00000411593.2
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr7_-_14029515 | 0.44 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chr14_-_72458326 | 0.44 |
ENST00000542853.1
|
AC005477.1
|
AC005477.1 |
| chr22_-_37172111 | 0.44 |
ENST00000417951.2
ENST00000430701.1 ENST00000433985.2 |
IFT27
|
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr10_-_100174900 | 0.43 |
ENST00000370575.4
|
PYROXD2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr11_+_74811578 | 0.43 |
ENST00000531713.1
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1 |
| chr20_-_23807358 | 0.43 |
ENST00000304725.2
|
CST2
|
cystatin SA |
| chr12_+_64846129 | 0.43 |
ENST00000540417.1
ENST00000539810.1 |
TBK1
|
TANK-binding kinase 1 |
| chr9_-_26892765 | 0.42 |
ENST00000520187.1
ENST00000333916.5 |
CAAP1
|
caspase activity and apoptosis inhibitor 1 |
| chr3_+_108308513 | 0.42 |
ENST00000361582.3
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr6_+_126112001 | 0.42 |
ENST00000392477.2
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr10_+_73975742 | 0.42 |
ENST00000299381.4
|
ANAPC16
|
anaphase promoting complex subunit 16 |
| chr7_-_14026063 | 0.42 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr13_+_24844819 | 0.42 |
ENST00000399949.2
|
SPATA13
|
spermatogenesis associated 13 |
| chr20_+_42187608 | 0.41 |
ENST00000373100.1
|
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr2_-_11484710 | 0.41 |
ENST00000315872.6
|
ROCK2
|
Rho-associated, coiled-coil containing protein kinase 2 |
| chr7_+_1084206 | 0.41 |
ENST00000444847.1
|
GPR146
|
G protein-coupled receptor 146 |
| chr4_+_105828492 | 0.41 |
ENST00000506148.1
|
RP11-556I14.1
|
RP11-556I14.1 |
| chr2_-_74648702 | 0.41 |
ENST00000518863.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr4_-_89442940 | 0.41 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr11_+_73000449 | 0.41 |
ENST00000535931.1
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr13_+_108921977 | 0.41 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr17_+_48450575 | 0.41 |
ENST00000338165.4
ENST00000393271.2 ENST00000511519.2 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
| chr10_+_115803650 | 0.40 |
ENST00000369295.2
|
ADRB1
|
adrenoceptor beta 1 |
| chr1_+_154966058 | 0.40 |
ENST00000392487.1
|
LENEP
|
lens epithelial protein |
| chr6_-_27858570 | 0.40 |
ENST00000359303.2
|
HIST1H3J
|
histone cluster 1, H3j |
| chr2_+_163200598 | 0.40 |
ENST00000437150.2
ENST00000453113.2 |
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr22_-_37172191 | 0.39 |
ENST00000340630.5
|
IFT27
|
intraflagellar transport 27 homolog (Chlamydomonas) |
| chrX_+_102883887 | 0.38 |
ENST00000372625.3
ENST00000372624.3 |
TCEAL1
|
transcription elongation factor A (SII)-like 1 |
| chr15_+_49913175 | 0.38 |
ENST00000403028.3
|
DTWD1
|
DTW domain containing 1 |
| chr3_-_119396193 | 0.38 |
ENST00000484810.1
ENST00000497116.1 ENST00000261070.2 |
COX17
|
COX17 cytochrome c oxidase copper chaperone |
| chr16_+_57496299 | 0.38 |
ENST00000219252.5
|
POLR2C
|
polymerase (RNA) II (DNA directed) polypeptide C, 33kDa |
| chr17_-_4689649 | 0.37 |
ENST00000441199.2
ENST00000416307.2 |
VMO1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chr8_-_145641864 | 0.37 |
ENST00000276833.5
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr8_-_145642267 | 0.37 |
ENST00000301305.3
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr17_-_79166176 | 0.37 |
ENST00000571292.1
|
AZI1
|
5-azacytidine induced 1 |
| chr4_-_15683230 | 0.37 |
ENST00000515679.1
|
FBXL5
|
F-box and leucine-rich repeat protein 5 |
| chr1_+_196788887 | 0.37 |
ENST00000320493.5
ENST00000367424.4 ENST00000367421.3 |
CFHR1
CFHR2
|
complement factor H-related 1 complement factor H-related 2 |
| chr5_+_140235469 | 0.36 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MAFF_MAFG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.5 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.4 | 1.6 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.4 | 1.2 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.3 | 1.4 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.3 | 2.2 | GO:0060356 | leucine import(GO:0060356) |
| 0.3 | 0.9 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.3 | 0.9 | GO:0090381 | regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.2 | 2.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 0.9 | GO:0048242 | regulation of epinephrine secretion(GO:0014060) negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) epinephrine secretion(GO:0048242) |
| 0.2 | 1.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.2 | 0.9 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.2 | 0.6 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.2 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.2 | 0.6 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.2 | 0.5 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) histone H3-K9 deacetylation(GO:1990619) |
| 0.2 | 0.5 | GO:0070409 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.2 | 0.5 | GO:0009968 | negative regulation of signal transduction(GO:0009968) |
| 0.2 | 0.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.8 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.2 | 1.3 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 1.1 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.6 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.4 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 0.5 | GO:1900155 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.1 | 2.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 2.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.5 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 0.4 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 0.9 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 5.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 2.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.5 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.9 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 0.7 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 0.3 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.7 | GO:1901523 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.1 | 0.8 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.8 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 2.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.8 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.6 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 1.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.7 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 0.4 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 13.0 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 1.8 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 0.5 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 0.8 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 1.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 0.2 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 0.2 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 1.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.1 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.1 | 0.2 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.1 | 0.2 | GO:1902869 | regulation of amacrine cell differentiation(GO:1902869) |
| 0.1 | 1.5 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.5 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.5 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 0.3 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 1.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 1.1 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.2 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.7 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.2 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.0 | 0.2 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.6 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0060981 | cell migration involved in coronary angiogenesis(GO:0060981) |
| 0.0 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.9 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.7 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.2 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 | 0.3 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.4 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.6 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 1.4 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.1 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.6 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 1.8 | GO:0030163 | protein catabolic process(GO:0030163) |
| 0.0 | 0.4 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 1.4 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.3 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.8 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.6 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.3 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 1.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:2001302 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.0 | 0.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 1.7 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 | 0.1 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.3 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.9 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.4 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0035377 | transepithelial water transport(GO:0035377) positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 1.3 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.4 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.4 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.5 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.5 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 2.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.2 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:1901340 | negative regulation of store-operated calcium channel activity(GO:1901340) |
| 0.0 | 0.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.3 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.5 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 0.3 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.3 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.0 | 0.1 | GO:0060136 | enucleate erythrocyte differentiation(GO:0043353) embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.3 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.8 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.0 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0017014 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.3 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:0046440 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.3 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 2.6 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.1 | GO:0046465 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.0 | 0.0 | GO:0031081 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 0.0 | 0.5 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.0 | 0.5 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.2 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.0 | 0.1 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.5 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.8 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.3 | 0.8 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.2 | 13.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.5 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 1.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.3 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 0.6 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.1 | 2.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.8 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 1.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 1.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 2.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.7 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 1.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 1.8 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.7 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.9 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.5 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.2 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.2 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 0.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 2.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 1.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.0 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 1.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 1.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) STAGA complex(GO:0030914) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 1.0 | 3.0 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.7 | 2.0 | GO:0047291 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.3 | 1.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.3 | 0.9 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.3 | 1.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.3 | 1.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.3 | 1.3 | GO:0047718 | androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.2 | 1.4 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.2 | 0.7 | GO:0097259 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.2 | 1.6 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.2 | 1.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 0.6 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.2 | 0.9 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 0.5 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.2 | 0.5 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.2 | 0.7 | GO:0047860 | diiodophenylpyruvate reductase activity(GO:0047860) |
| 0.2 | 14.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.2 | 3.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 0.8 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.9 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.1 | 1.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.5 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 0.5 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.4 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 2.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 1.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 2.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.5 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.1 | 1.0 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 2.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.5 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.2 | GO:0031768 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.1 | 0.5 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.7 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.7 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 0.7 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.2 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 0.9 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.1 | 1.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.9 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 1.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.5 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.7 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 1.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0016826 | N-sulfoglucosamine sulfohydrolase activity(GO:0016250) hydrolase activity, acting on acid sulfur-nitrogen bonds(GO:0016826) |
| 0.0 | 0.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.9 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 1.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 1.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 1.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.5 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 1.0 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 1.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 2.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.0 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.0 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.0 | 0.1 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.6 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.0 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.9 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.0 | GO:0004423 | iduronate-2-sulfatase activity(GO:0004423) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0003690 | double-stranded DNA binding(GO:0003690) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 2.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 13.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 2.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 2.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.8 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 2.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.8 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 1.1 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 2.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.9 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |