Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
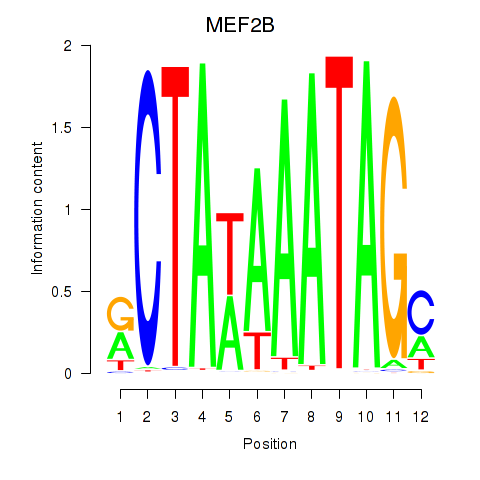
Results for MEF2B
Z-value: 0.43
Transcription factors associated with MEF2B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEF2B
|
ENSG00000213999.11 | myocyte enhancer factor 2B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEF2B | hg19_v2_chr19_-_19281054_19281098 | 0.51 | 2.3e-02 | Click! |
Activity profile of MEF2B motif
Sorted Z-values of MEF2B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_52445191 | 6.19 |
ENST00000243050.1
ENST00000394825.1 ENST00000550763.1 ENST00000394824.2 ENST00000548232.1 ENST00000562373.1 |
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr12_+_52430894 | 1.90 |
ENST00000546842.1
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr20_+_10199468 | 1.78 |
ENST00000254976.2
ENST00000304886.2 |
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr4_+_41362796 | 1.59 |
ENST00000508501.1
ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr11_+_71934962 | 1.29 |
ENST00000543234.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr22_-_51017084 | 1.10 |
ENST00000360719.2
ENST00000457250.1 ENST00000440709.1 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr9_+_97766409 | 1.02 |
ENST00000425634.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr22_-_51016846 | 1.01 |
ENST00000312108.7
ENST00000395650.2 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr6_-_33160231 | 0.94 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr1_+_156095951 | 0.92 |
ENST00000448611.2
ENST00000368297.1 |
LMNA
|
lamin A/C |
| chr1_-_204329013 | 0.91 |
ENST00000272203.3
ENST00000414478.1 |
PLEKHA6
|
pleckstrin homology domain containing, family A member 6 |
| chr5_+_155753745 | 0.89 |
ENST00000435422.3
ENST00000337851.4 ENST00000447401.1 |
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| chr17_-_42200996 | 0.88 |
ENST00000587135.1
ENST00000225983.6 ENST00000393622.2 ENST00000588703.1 |
HDAC5
|
histone deacetylase 5 |
| chr2_-_165424973 | 0.86 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr2_-_157198860 | 0.86 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr9_-_74980113 | 0.84 |
ENST00000376962.5
ENST00000376960.4 ENST00000237937.3 |
ZFAND5
|
zinc finger, AN1-type domain 5 |
| chr2_-_86564776 | 0.83 |
ENST00000165698.5
ENST00000541910.1 ENST00000535845.1 |
REEP1
|
receptor accessory protein 1 |
| chr16_-_88717482 | 0.82 |
ENST00000261623.3
|
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr1_+_156096336 | 0.80 |
ENST00000504687.1
ENST00000473598.2 |
LMNA
|
lamin A/C |
| chr3_+_8543561 | 0.79 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr20_+_10199566 | 0.78 |
ENST00000430336.1
|
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr7_-_16872932 | 0.77 |
ENST00000419572.2
ENST00000412973.1 |
AGR2
|
anterior gradient 2 |
| chr12_+_52431016 | 0.71 |
ENST00000553200.1
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr17_-_42200958 | 0.70 |
ENST00000336057.5
|
HDAC5
|
histone deacetylase 5 |
| chr12_-_49393092 | 0.70 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr3_+_8543393 | 0.70 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr3_-_52486841 | 0.65 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr12_+_54384370 | 0.62 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chr4_+_41362615 | 0.60 |
ENST00000509638.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr3_+_8543533 | 0.59 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr22_-_30642782 | 0.56 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr9_+_100263912 | 0.56 |
ENST00000259365.4
|
TMOD1
|
tropomodulin 1 |
| chr3_+_49057876 | 0.56 |
ENST00000326912.4
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr17_-_48785216 | 0.55 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr8_+_76452097 | 0.53 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr16_-_88717423 | 0.49 |
ENST00000568278.1
ENST00000569359.1 ENST00000567174.1 |
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr2_+_33661382 | 0.48 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr2_+_233497931 | 0.48 |
ENST00000264059.3
|
EFHD1
|
EF-hand domain family, member D1 |
| chr1_+_212782012 | 0.47 |
ENST00000341491.4
ENST00000366985.1 |
ATF3
|
activating transcription factor 3 |
| chr17_-_42188598 | 0.45 |
ENST00000591714.1
|
HDAC5
|
histone deacetylase 5 |
| chr2_-_190044480 | 0.45 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr22_-_30642728 | 0.45 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr7_-_37488834 | 0.45 |
ENST00000310758.4
|
ELMO1
|
engulfment and cell motility 1 |
| chr11_+_7597639 | 0.43 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr2_-_86564740 | 0.43 |
ENST00000540790.1
ENST00000428491.1 |
REEP1
|
receptor accessory protein 1 |
| chr3_+_179370517 | 0.42 |
ENST00000263966.3
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr5_-_16509101 | 0.42 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr19_-_40324767 | 0.42 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chrX_+_70521584 | 0.40 |
ENST00000373829.3
ENST00000538820.1 |
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2 |
| chr19_-_40324255 | 0.39 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr3_-_137834436 | 0.39 |
ENST00000327532.2
ENST00000467030.1 |
DZIP1L
|
DAZ interacting zinc finger protein 1-like |
| chr17_-_5322786 | 0.39 |
ENST00000225696.4
|
NUP88
|
nucleoporin 88kDa |
| chr11_-_18034701 | 0.38 |
ENST00000265965.5
|
SERGEF
|
secretion regulating guanine nucleotide exchange factor |
| chr17_-_34417479 | 0.37 |
ENST00000225245.5
|
CCL3
|
chemokine (C-C motif) ligand 3 |
| chr6_+_44194762 | 0.36 |
ENST00000371708.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr7_-_121944491 | 0.33 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr1_-_150693318 | 0.32 |
ENST00000442853.1
ENST00000368995.4 ENST00000368993.2 ENST00000361824.2 ENST00000322343.7 |
HORMAD1
|
HORMA domain containing 1 |
| chr2_-_86564696 | 0.32 |
ENST00000437769.1
|
REEP1
|
receptor accessory protein 1 |
| chr3_-_192445289 | 0.32 |
ENST00000430714.1
ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12
|
fibroblast growth factor 12 |
| chr7_-_37488777 | 0.32 |
ENST00000445322.1
ENST00000448602.1 |
ELMO1
|
engulfment and cell motility 1 |
| chr12_-_120189900 | 0.31 |
ENST00000546026.1
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr8_-_62602327 | 0.31 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr17_-_1389228 | 0.30 |
ENST00000438665.2
|
MYO1C
|
myosin IC |
| chr19_-_16008880 | 0.30 |
ENST00000011989.7
ENST00000221700.6 |
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr11_+_47293795 | 0.29 |
ENST00000422579.1
|
MADD
|
MAP-kinase activating death domain |
| chr18_+_12407895 | 0.29 |
ENST00000590956.1
ENST00000336990.4 ENST00000440960.1 ENST00000588729.1 |
SLMO1
|
slowmo homolog 1 (Drosophila) |
| chr4_+_120056939 | 0.27 |
ENST00000307128.5
|
MYOZ2
|
myozenin 2 |
| chr20_+_11898507 | 0.27 |
ENST00000378226.2
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr17_+_42248063 | 0.27 |
ENST00000293414.1
|
ASB16
|
ankyrin repeat and SOCS box containing 16 |
| chr9_-_85882145 | 0.26 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr9_-_16728161 | 0.24 |
ENST00000603713.1
ENST00000603313.1 |
BNC2
|
basonuclin 2 |
| chr11_-_47374246 | 0.23 |
ENST00000545968.1
ENST00000399249.2 ENST00000256993.4 |
MYBPC3
|
myosin binding protein C, cardiac |
| chr1_+_145524891 | 0.20 |
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10 |
| chr5_-_11903337 | 0.20 |
ENST00000502551.1
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr1_+_145525015 | 0.19 |
ENST00000539363.1
ENST00000538811.1 |
ITGA10
|
integrin, alpha 10 |
| chr8_-_27115903 | 0.19 |
ENST00000350889.4
ENST00000519997.1 ENST00000519614.1 ENST00000522908.1 ENST00000265770.7 |
STMN4
|
stathmin-like 4 |
| chr1_-_150693305 | 0.19 |
ENST00000368987.1
|
HORMAD1
|
HORMA domain containing 1 |
| chr20_+_32581525 | 0.18 |
ENST00000246194.3
ENST00000413297.1 |
RALY
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr5_-_111091948 | 0.18 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr17_+_20059302 | 0.18 |
ENST00000395530.2
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr20_-_62582475 | 0.17 |
ENST00000369908.5
|
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr14_+_76776957 | 0.17 |
ENST00000512784.1
|
ESRRB
|
estrogen-related receptor beta |
| chr17_-_1389419 | 0.16 |
ENST00000575158.1
|
MYO1C
|
myosin IC |
| chr16_+_50313426 | 0.16 |
ENST00000569265.1
|
ADCY7
|
adenylate cyclase 7 |
| chr2_+_170366203 | 0.16 |
ENST00000284669.1
|
KLHL41
|
kelch-like family member 41 |
| chrX_-_15332665 | 0.16 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr4_+_8321882 | 0.16 |
ENST00000509453.1
ENST00000503186.1 |
RP11-774O3.2
RP11-774O3.1
|
RP11-774O3.2 RP11-774O3.1 |
| chr11_+_7598239 | 0.15 |
ENST00000525597.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr8_-_72268889 | 0.14 |
ENST00000388742.4
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr16_+_12059091 | 0.14 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr5_-_11903475 | 0.14 |
ENST00000508761.1
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr8_-_27115931 | 0.13 |
ENST00000523048.1
|
STMN4
|
stathmin-like 4 |
| chr14_+_101359265 | 0.13 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr2_-_218766698 | 0.13 |
ENST00000439083.1
|
TNS1
|
tensin 1 |
| chr5_+_132009675 | 0.13 |
ENST00000231449.2
ENST00000350025.2 |
IL4
|
interleukin 4 |
| chr6_+_123038689 | 0.12 |
ENST00000354275.2
ENST00000368446.1 |
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr6_-_123957942 | 0.11 |
ENST00000398178.3
|
TRDN
|
triadin |
| chr8_-_72268968 | 0.11 |
ENST00000388740.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr18_-_35145981 | 0.11 |
ENST00000420428.2
ENST00000412753.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr7_-_37488547 | 0.11 |
ENST00000453399.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr6_-_123958051 | 0.10 |
ENST00000546248.1
|
TRDN
|
triadin |
| chr1_+_74701062 | 0.10 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr10_-_97200772 | 0.10 |
ENST00000371241.1
ENST00000354106.3 ENST00000371239.1 ENST00000361941.3 ENST00000277982.5 ENST00000371245.3 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr18_-_3220106 | 0.10 |
ENST00000356443.4
ENST00000400569.3 |
MYOM1
|
myomesin 1 |
| chr3_-_39234074 | 0.09 |
ENST00000340369.3
ENST00000421646.1 ENST00000396251.1 |
XIRP1
|
xin actin-binding repeat containing 1 |
| chr3_+_54157480 | 0.09 |
ENST00000490478.1
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr18_-_3219847 | 0.08 |
ENST00000261606.7
|
MYOM1
|
myomesin 1 |
| chr11_-_47400062 | 0.08 |
ENST00000533030.1
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr5_-_66492562 | 0.08 |
ENST00000256447.4
|
CD180
|
CD180 molecule |
| chr11_-_104480019 | 0.08 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chrX_-_138304939 | 0.08 |
ENST00000448673.1
|
FGF13
|
fibroblast growth factor 13 |
| chr17_-_27467418 | 0.08 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr10_+_71561630 | 0.08 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr4_-_185303418 | 0.07 |
ENST00000610223.1
ENST00000608785.1 |
RP11-290F5.1
|
RP11-290F5.1 |
| chr7_+_86781916 | 0.07 |
ENST00000579592.1
ENST00000434534.1 |
DMTF1
|
cyclin D binding myb-like transcription factor 1 |
| chr7_+_75931861 | 0.07 |
ENST00000248553.6
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr3_+_46924829 | 0.07 |
ENST00000313049.5
|
PTH1R
|
parathyroid hormone 1 receptor |
| chr10_-_75415825 | 0.07 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr1_+_84630645 | 0.07 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr12_-_99548270 | 0.06 |
ENST00000546568.1
ENST00000332712.7 ENST00000546960.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr14_-_70546897 | 0.06 |
ENST00000394330.2
ENST00000533541.1 ENST00000216568.7 |
SLC8A3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chrX_-_142722897 | 0.06 |
ENST00000338017.4
|
SLITRK4
|
SLIT and NTRK-like family, member 4 |
| chr10_+_71561704 | 0.06 |
ENST00000520267.1
|
COL13A1
|
collagen, type XIII, alpha 1 |
| chr12_+_15699286 | 0.05 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr11_-_47400032 | 0.05 |
ENST00000533968.1
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr7_-_15726296 | 0.05 |
ENST00000262041.5
|
MEOX2
|
mesenchyme homeobox 2 |
| chr14_-_102704783 | 0.05 |
ENST00000522534.1
|
MOK
|
MOK protein kinase |
| chr9_+_116327326 | 0.05 |
ENST00000342620.5
|
RGS3
|
regulator of G-protein signaling 3 |
| chr2_+_33683109 | 0.05 |
ENST00000437184.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr10_-_69455873 | 0.05 |
ENST00000433211.2
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr11_-_84634447 | 0.05 |
ENST00000532653.1
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr11_-_47400078 | 0.04 |
ENST00000378538.3
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr18_+_3252206 | 0.04 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr3_-_11610255 | 0.04 |
ENST00000424529.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr13_+_39261224 | 0.04 |
ENST00000280481.7
|
FREM2
|
FRAS1 related extracellular matrix protein 2 |
| chr2_-_145188137 | 0.04 |
ENST00000440875.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr1_+_84630367 | 0.04 |
ENST00000370680.1
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr7_+_30951461 | 0.03 |
ENST00000311813.4
|
AQP1
|
aquaporin 1 (Colton blood group) |
| chr12_-_5352315 | 0.03 |
ENST00000536518.1
|
RP11-319E16.1
|
RP11-319E16.1 |
| chr16_-_56223480 | 0.03 |
ENST00000565155.1
|
RP11-461O7.1
|
RP11-461O7.1 |
| chr9_-_102582155 | 0.03 |
ENST00000427039.1
|
RP11-554F20.1
|
RP11-554F20.1 |
| chr11_-_84634217 | 0.03 |
ENST00000524982.1
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr2_+_168043793 | 0.03 |
ENST00000409273.1
ENST00000409605.1 |
XIRP2
|
xin actin-binding repeat containing 2 |
| chr10_+_71561649 | 0.03 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr14_-_22005343 | 0.03 |
ENST00000327430.3
|
SALL2
|
spalt-like transcription factor 2 |
| chr20_+_44657845 | 0.02 |
ENST00000243964.3
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr11_-_47399942 | 0.02 |
ENST00000227163.4
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr12_-_371994 | 0.02 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr4_+_37892682 | 0.02 |
ENST00000508802.1
ENST00000261439.4 ENST00000402522.1 |
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr11_+_75110530 | 0.02 |
ENST00000531188.1
ENST00000530164.1 ENST00000422465.2 ENST00000278572.6 ENST00000534440.1 ENST00000527446.1 ENST00000526608.1 ENST00000527273.1 ENST00000524851.1 |
RPS3
|
ribosomal protein S3 |
| chr16_+_31225337 | 0.02 |
ENST00000322122.3
|
TRIM72
|
tripartite motif containing 72 |
| chr7_+_86781847 | 0.01 |
ENST00000432366.2
ENST00000423590.2 ENST00000394703.5 |
DMTF1
|
cyclin D binding myb-like transcription factor 1 |
| chr20_+_61147651 | 0.01 |
ENST00000370527.3
ENST00000370524.2 |
C20orf166
|
chromosome 20 open reading frame 166 |
| chrX_-_63450480 | 0.01 |
ENST00000362002.2
|
ASB12
|
ankyrin repeat and SOCS box containing 12 |
| chrX_-_11284095 | 0.01 |
ENST00000303025.6
ENST00000534860.1 |
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr8_+_11351876 | 0.01 |
ENST00000529894.1
|
BLK
|
B lymphoid tyrosine kinase |
| chr10_-_31146615 | 0.01 |
ENST00000444692.2
|
ZNF438
|
zinc finger protein 438 |
| chr6_+_139094657 | 0.00 |
ENST00000332797.6
|
CCDC28A
|
coiled-coil domain containing 28A |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.7 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.6 | 2.6 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.3 | 1.7 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.3 | 1.3 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.2 | 1.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.2 | 0.9 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 0.5 | GO:0060629 | regulation of homologous chromosome segregation(GO:0060629) |
| 0.2 | 0.6 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.2 | 0.5 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.2 | 0.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.4 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.8 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.1 | 0.3 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 2.1 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.3 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 2.1 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.1 | 0.2 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.1 | 0.4 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 1.6 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.2 | GO:1901846 | positive regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901846) |
| 0.1 | 2.1 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.4 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 1.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.8 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.9 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.2 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.9 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.0 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.6 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.2 | 1.7 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 0.5 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.2 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.5 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.4 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.4 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 0.9 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 1.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 1.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 8.0 | GO:0031965 | nuclear membrane(GO:0031965) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.3 | 1.6 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.3 | 2.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.2 | 2.0 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 0.3 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.3 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 10.3 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 0.9 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 1.3 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 0.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.8 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 1.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.4 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 2.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.4 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.0 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 2.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 2.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.8 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 2.6 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 2.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.9 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |