Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for MEF2C
Z-value: 0.43
Transcription factors associated with MEF2C
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEF2C
|
ENSG00000081189.9 | myocyte enhancer factor 2C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEF2C | hg19_v2_chr5_-_88178964_88179012 | 0.80 | 1.9e-05 | Click! |
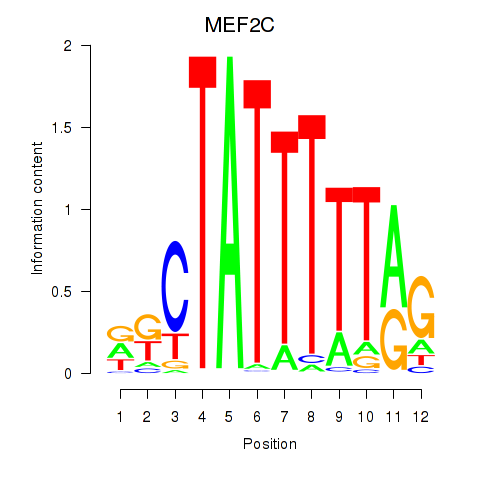
Activity profile of MEF2C motif
Sorted Z-values of MEF2C motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_143226979 | 1.85 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr4_-_143227088 | 1.79 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr8_-_49834299 | 1.78 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr8_-_49833978 | 1.70 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr1_-_209975494 | 1.46 |
ENST00000456314.1
|
IRF6
|
interferon regulatory factor 6 |
| chr3_+_99357319 | 1.42 |
ENST00000452013.1
ENST00000261037.3 ENST00000273342.4 |
COL8A1
|
collagen, type VIII, alpha 1 |
| chr18_+_29027696 | 1.35 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr1_+_68150744 | 1.22 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr3_-_176914191 | 1.22 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr15_+_78558523 | 1.14 |
ENST00000446172.2
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr7_+_18535321 | 1.08 |
ENST00000413380.1
ENST00000430454.1 |
HDAC9
|
histone deacetylase 9 |
| chr3_+_135741576 | 1.07 |
ENST00000334546.2
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr14_-_23652849 | 1.01 |
ENST00000316902.7
ENST00000469263.1 ENST00000525062.1 ENST00000524758.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr10_-_3827371 | 1.01 |
ENST00000469435.1
|
KLF6
|
Kruppel-like factor 6 |
| chr7_+_18535346 | 1.00 |
ENST00000405010.3
ENST00000406451.4 ENST00000428307.2 |
HDAC9
|
histone deacetylase 9 |
| chr4_-_143767428 | 0.98 |
ENST00000513000.1
ENST00000509777.1 ENST00000503927.1 |
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr17_+_80186908 | 0.97 |
ENST00000582743.1
ENST00000578684.1 ENST00000577650.1 ENST00000582715.1 |
SLC16A3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr14_-_61748550 | 0.94 |
ENST00000555868.1
|
TMEM30B
|
transmembrane protein 30B |
| chr2_-_151344172 | 0.91 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr6_+_15401075 | 0.87 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr2_-_208031542 | 0.82 |
ENST00000423015.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr10_-_3827417 | 0.77 |
ENST00000497571.1
ENST00000542957.1 |
KLF6
|
Kruppel-like factor 6 |
| chr8_-_139926236 | 0.76 |
ENST00000303045.6
ENST00000435777.1 |
COL22A1
|
collagen, type XXII, alpha 1 |
| chr4_+_89514402 | 0.76 |
ENST00000426683.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr1_+_230193521 | 0.74 |
ENST00000543760.1
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr21_+_39628852 | 0.72 |
ENST00000398938.2
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr2_-_208030886 | 0.70 |
ENST00000426163.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr4_+_160203650 | 0.68 |
ENST00000514565.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr7_+_18126557 | 0.67 |
ENST00000417496.2
|
HDAC9
|
histone deacetylase 9 |
| chr4_+_89514516 | 0.65 |
ENST00000452979.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr7_+_18535893 | 0.64 |
ENST00000432645.2
ENST00000441542.2 |
HDAC9
|
histone deacetylase 9 |
| chrX_+_131157609 | 0.63 |
ENST00000496850.1
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr2_+_148778570 | 0.62 |
ENST00000407073.1
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr15_+_42651691 | 0.62 |
ENST00000357568.3
ENST00000349748.3 ENST00000318023.7 ENST00000397163.3 |
CAPN3
|
calpain 3, (p94) |
| chr5_-_137475071 | 0.62 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr18_+_9708162 | 0.61 |
ENST00000578921.1
|
RAB31
|
RAB31, member RAS oncogene family |
| chr20_+_22034809 | 0.61 |
ENST00000449427.1
|
RP11-125P18.1
|
RP11-125P18.1 |
| chr3_-_48598547 | 0.60 |
ENST00000536104.1
ENST00000452531.1 |
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr3_+_138067521 | 0.58 |
ENST00000494949.1
|
MRAS
|
muscle RAS oncogene homolog |
| chr21_+_39628655 | 0.58 |
ENST00000398925.1
ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr20_+_58251716 | 0.57 |
ENST00000355648.4
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr7_+_18536090 | 0.57 |
ENST00000441986.1
|
HDAC9
|
histone deacetylase 9 |
| chr22_+_32340447 | 0.57 |
ENST00000248975.5
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr1_-_230513367 | 0.56 |
ENST00000321327.2
ENST00000525115.1 |
PGBD5
|
piggyBac transposable element derived 5 |
| chr8_+_104831472 | 0.56 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr22_+_32340481 | 0.56 |
ENST00000397492.1
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr3_-_176914238 | 0.56 |
ENST00000430069.1
ENST00000428970.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr9_+_127054254 | 0.55 |
ENST00000422297.1
|
NEK6
|
NIMA-related kinase 6 |
| chr15_+_68582544 | 0.54 |
ENST00000566008.1
|
FEM1B
|
fem-1 homolog b (C. elegans) |
| chr13_+_76378357 | 0.52 |
ENST00000489941.2
ENST00000525373.1 |
LMO7
|
LIM domain 7 |
| chr4_+_169418255 | 0.52 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr3_+_138067314 | 0.52 |
ENST00000423968.2
|
MRAS
|
muscle RAS oncogene homolog |
| chr2_+_220144168 | 0.50 |
ENST00000392087.2
ENST00000442681.1 ENST00000439026.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr12_+_14561422 | 0.49 |
ENST00000541056.1
|
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr3_+_138067666 | 0.48 |
ENST00000475711.1
ENST00000464896.1 |
MRAS
|
muscle RAS oncogene homolog |
| chr22_+_41487711 | 0.48 |
ENST00000263253.7
|
EP300
|
E1A binding protein p300 |
| chr3_-_112360116 | 0.48 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chr1_+_201617450 | 0.46 |
ENST00000295624.6
ENST00000367297.4 ENST00000367300.3 |
NAV1
|
neuron navigator 1 |
| chr3_+_141106458 | 0.46 |
ENST00000509883.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr13_+_76334498 | 0.46 |
ENST00000534657.1
|
LMO7
|
LIM domain 7 |
| chr15_+_40531243 | 0.45 |
ENST00000558055.1
ENST00000455577.2 |
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr2_+_182850743 | 0.45 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr8_+_107282389 | 0.45 |
ENST00000577661.1
ENST00000445937.1 |
RP11-395G23.3
OXR1
|
RP11-395G23.3 oxidation resistance 1 |
| chr3_-_149510553 | 0.45 |
ENST00000462519.2
ENST00000446160.1 ENST00000383050.3 |
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr16_+_14280742 | 0.45 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr3_-_38071122 | 0.45 |
ENST00000334661.4
|
PLCD1
|
phospholipase C, delta 1 |
| chr2_+_220299547 | 0.44 |
ENST00000312358.7
|
SPEG
|
SPEG complex locus |
| chr2_-_208030647 | 0.43 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr1_-_201346761 | 0.42 |
ENST00000455702.1
ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2
|
troponin T type 2 (cardiac) |
| chr11_-_85780853 | 0.42 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr19_+_37096194 | 0.41 |
ENST00000460670.1
ENST00000292928.2 ENST00000439428.1 |
ZNF382
|
zinc finger protein 382 |
| chr1_-_201390846 | 0.41 |
ENST00000367312.1
ENST00000555340.2 ENST00000361379.4 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr15_+_40531621 | 0.41 |
ENST00000560346.1
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr2_+_220144052 | 0.41 |
ENST00000425450.1
ENST00000392086.4 ENST00000421532.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr1_-_31845914 | 0.41 |
ENST00000373713.2
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr13_+_76378407 | 0.41 |
ENST00000447038.1
|
LMO7
|
LIM domain 7 |
| chr5_-_142065223 | 0.40 |
ENST00000378046.1
|
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr4_+_160025300 | 0.40 |
ENST00000505478.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr13_-_30881134 | 0.39 |
ENST00000380617.3
ENST00000441394.1 |
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr17_+_7461580 | 0.39 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr1_+_16010779 | 0.39 |
ENST00000375799.3
ENST00000375793.2 |
PLEKHM2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr13_+_76378305 | 0.39 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr5_+_96077888 | 0.38 |
ENST00000509259.1
ENST00000503828.1 |
CAST
|
calpastatin |
| chr4_-_186697044 | 0.38 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_-_115910630 | 0.38 |
ENST00000343348.6
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr8_+_107282368 | 0.38 |
ENST00000521369.2
|
RP11-395G23.3
|
RP11-395G23.3 |
| chr1_-_226187013 | 0.37 |
ENST00000272091.7
|
SDE2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr11_+_69924639 | 0.37 |
ENST00000538023.1
ENST00000398543.2 |
ANO1
|
anoctamin 1, calcium activated chloride channel |
| chr11_+_1860200 | 0.37 |
ENST00000381911.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr4_-_25865159 | 0.37 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr7_+_90032821 | 0.36 |
ENST00000427904.1
|
CLDN12
|
claudin 12 |
| chr1_+_201617264 | 0.36 |
ENST00000367296.4
|
NAV1
|
neuron navigator 1 |
| chr3_-_168864315 | 0.36 |
ENST00000475754.1
ENST00000484519.1 |
MECOM
|
MDS1 and EVI1 complex locus |
| chr8_-_73793975 | 0.36 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr10_-_29923893 | 0.36 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr1_+_90098606 | 0.35 |
ENST00000370454.4
|
LRRC8C
|
leucine rich repeat containing 8 family, member C |
| chr12_+_53846594 | 0.35 |
ENST00000550192.1
|
PCBP2
|
poly(rC) binding protein 2 |
| chrX_-_50386648 | 0.35 |
ENST00000460112.3
|
SHROOM4
|
shroom family member 4 |
| chr3_+_186739636 | 0.35 |
ENST00000440338.1
ENST00000448044.1 |
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr10_-_62060232 | 0.35 |
ENST00000503925.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr14_-_21493123 | 0.34 |
ENST00000556147.1
ENST00000554489.1 ENST00000555657.1 ENST00000557274.1 ENST00000555158.1 ENST00000554833.1 ENST00000555384.1 ENST00000556420.1 ENST00000554893.1 ENST00000553503.1 ENST00000555733.1 ENST00000553867.1 ENST00000397856.3 ENST00000397855.3 ENST00000556008.1 ENST00000557182.1 ENST00000554483.1 ENST00000556688.1 ENST00000397853.3 ENST00000556329.2 ENST00000554143.1 ENST00000397851.2 ENST00000555142.1 ENST00000557676.1 ENST00000556924.1 |
NDRG2
|
NDRG family member 2 |
| chr9_-_110251836 | 0.33 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr5_+_191592 | 0.33 |
ENST00000328278.3
|
LRRC14B
|
leucine rich repeat containing 14B |
| chr22_+_19706958 | 0.33 |
ENST00000395109.2
|
SEPT5
|
septin 5 |
| chr4_-_57547870 | 0.32 |
ENST00000381260.3
ENST00000420433.1 ENST00000554144.1 ENST00000557328.1 |
HOPX
|
HOP homeobox |
| chr8_-_72268721 | 0.32 |
ENST00000419131.1
ENST00000388743.2 |
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr16_-_18923035 | 0.32 |
ENST00000563836.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr10_-_61495760 | 0.32 |
ENST00000395347.1
|
SLC16A9
|
solute carrier family 16, member 9 |
| chr2_+_220143989 | 0.32 |
ENST00000336576.5
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr5_+_138609782 | 0.31 |
ENST00000361059.2
ENST00000514694.1 ENST00000504203.1 ENST00000502929.1 ENST00000394800.2 ENST00000509644.1 ENST00000505016.1 |
MATR3
|
matrin 3 |
| chr17_+_17685422 | 0.30 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chrX_-_15683147 | 0.29 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr10_+_124134201 | 0.29 |
ENST00000368990.3
ENST00000368988.1 ENST00000368989.2 ENST00000463663.2 |
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr8_-_95565673 | 0.29 |
ENST00000437199.1
ENST00000297591.5 ENST00000421249.2 |
KIAA1429
|
KIAA1429 |
| chr11_+_128563652 | 0.29 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chrX_-_109683446 | 0.29 |
ENST00000372057.1
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr13_-_30880979 | 0.29 |
ENST00000414289.1
|
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr5_-_142065612 | 0.29 |
ENST00000360966.5
ENST00000411960.1 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr21_+_39628780 | 0.28 |
ENST00000417042.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr14_+_58797974 | 0.28 |
ENST00000417477.2
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr11_+_65266507 | 0.28 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chrX_+_135279179 | 0.28 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr12_+_53846612 | 0.28 |
ENST00000551104.1
|
PCBP2
|
poly(rC) binding protein 2 |
| chr8_+_132952112 | 0.27 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr16_+_14280564 | 0.27 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr2_+_136343943 | 0.26 |
ENST00000409606.1
|
R3HDM1
|
R3H domain containing 1 |
| chr1_+_160051319 | 0.26 |
ENST00000368088.3
|
KCNJ9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr14_+_56046914 | 0.26 |
ENST00000413890.2
ENST00000395309.3 ENST00000554567.1 ENST00000555498.1 |
KTN1
|
kinectin 1 (kinesin receptor) |
| chr1_+_24018269 | 0.26 |
ENST00000374550.3
|
RPL11
|
ribosomal protein L11 |
| chr18_+_55862873 | 0.25 |
ENST00000588494.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr10_+_102758105 | 0.25 |
ENST00000429732.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr5_+_53751445 | 0.25 |
ENST00000302005.1
|
HSPB3
|
heat shock 27kDa protein 3 |
| chr14_-_74462922 | 0.25 |
ENST00000553284.1
|
ENTPD5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr9_+_127054217 | 0.24 |
ENST00000394199.2
ENST00000546191.1 |
NEK6
|
NIMA-related kinase 6 |
| chr8_-_70745575 | 0.24 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr13_+_97928395 | 0.24 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr10_-_14880002 | 0.23 |
ENST00000465530.1
|
CDNF
|
cerebral dopamine neurotrophic factor |
| chr4_-_186696561 | 0.23 |
ENST00000445115.1
ENST00000451701.1 ENST00000457247.1 ENST00000435480.1 ENST00000425679.1 ENST00000457934.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr19_+_6739662 | 0.23 |
ENST00000313285.8
ENST00000313244.9 ENST00000596758.1 |
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr7_-_143956815 | 0.23 |
ENST00000493325.1
|
OR2A7
|
olfactory receptor, family 2, subfamily A, member 7 |
| chr11_-_19223523 | 0.23 |
ENST00000265968.3
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr5_+_173316341 | 0.23 |
ENST00000520867.1
ENST00000334035.5 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr1_-_59249732 | 0.23 |
ENST00000371222.2
|
JUN
|
jun proto-oncogene |
| chr11_+_19799327 | 0.23 |
ENST00000540292.1
|
NAV2
|
neuron navigator 2 |
| chr4_+_100495864 | 0.22 |
ENST00000265517.5
ENST00000422897.2 |
MTTP
|
microsomal triglyceride transfer protein |
| chr10_-_61900762 | 0.22 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr3_+_159570722 | 0.22 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr2_-_220117867 | 0.21 |
ENST00000456818.1
ENST00000447205.1 |
TUBA4A
|
tubulin, alpha 4a |
| chr9_+_127023704 | 0.21 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr7_-_151217166 | 0.21 |
ENST00000496004.1
|
RHEB
|
Ras homolog enriched in brain |
| chr14_-_94421923 | 0.21 |
ENST00000555507.1
|
ASB2
|
ankyrin repeat and SOCS box containing 2 |
| chr3_+_154797877 | 0.21 |
ENST00000462745.1
ENST00000493237.1 |
MME
|
membrane metallo-endopeptidase |
| chr1_-_156460391 | 0.21 |
ENST00000360595.3
|
MEF2D
|
myocyte enhancer factor 2D |
| chr11_+_1860682 | 0.21 |
ENST00000381906.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr17_+_7758374 | 0.21 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr14_+_56046990 | 0.20 |
ENST00000438792.2
ENST00000395314.3 ENST00000395308.1 |
KTN1
|
kinectin 1 (kinesin receptor) |
| chrX_-_131353461 | 0.20 |
ENST00000370874.1
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr2_+_179184955 | 0.20 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr8_+_104831554 | 0.20 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr10_+_63661053 | 0.20 |
ENST00000279873.7
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr13_+_111766897 | 0.19 |
ENST00000317133.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr1_-_116383322 | 0.19 |
ENST00000429731.1
|
NHLH2
|
nescient helix loop helix 2 |
| chr1_+_114473350 | 0.19 |
ENST00000503968.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr11_+_69924679 | 0.19 |
ENST00000531604.1
|
ANO1
|
anoctamin 1, calcium activated chloride channel |
| chr7_-_124569864 | 0.19 |
ENST00000609702.1
|
POT1
|
protection of telomeres 1 |
| chr9_-_21995249 | 0.19 |
ENST00000494262.1
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A |
| chr18_+_32402321 | 0.19 |
ENST00000587723.1
|
DTNA
|
dystrobrevin, alpha |
| chr7_-_124569991 | 0.19 |
ENST00000446993.1
ENST00000357628.3 ENST00000393329.1 |
POT1
|
protection of telomeres 1 |
| chr22_-_31328881 | 0.18 |
ENST00000445980.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr1_-_154164534 | 0.18 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr1_+_92632542 | 0.18 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr18_+_42260059 | 0.18 |
ENST00000426838.4
|
SETBP1
|
SET binding protein 1 |
| chr7_-_105332084 | 0.18 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr13_+_76334795 | 0.17 |
ENST00000526202.1
ENST00000465261.2 |
LMO7
|
LIM domain 7 |
| chr8_-_71157595 | 0.17 |
ENST00000519724.1
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr1_+_183605222 | 0.17 |
ENST00000536277.1
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr1_-_152131703 | 0.17 |
ENST00000316073.3
|
RPTN
|
repetin |
| chr1_+_104104379 | 0.17 |
ENST00000435302.1
|
AMY2B
|
amylase, alpha 2B (pancreatic) |
| chr11_+_1940925 | 0.16 |
ENST00000453458.1
ENST00000381557.2 ENST00000381589.3 ENST00000381579.3 ENST00000381563.4 ENST00000344578.4 |
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr4_-_170192000 | 0.16 |
ENST00000502315.1
|
SH3RF1
|
SH3 domain containing ring finger 1 |
| chr3_+_4535025 | 0.16 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr11_+_1860832 | 0.16 |
ENST00000252898.7
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr1_+_41447609 | 0.16 |
ENST00000543104.1
|
CTPS1
|
CTP synthase 1 |
| chr8_+_104831440 | 0.16 |
ENST00000515551.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr1_-_204436344 | 0.16 |
ENST00000367184.2
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr4_-_186696425 | 0.16 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_-_214017151 | 0.16 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr7_-_55583740 | 0.15 |
ENST00000453256.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr5_-_131347501 | 0.15 |
ENST00000543479.1
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr8_+_22225041 | 0.15 |
ENST00000289952.5
ENST00000524285.1 |
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr4_-_57547454 | 0.15 |
ENST00000556376.2
|
HOPX
|
HOP homeobox |
| chr15_+_100106126 | 0.15 |
ENST00000558812.1
ENST00000338042.6 |
MEF2A
|
myocyte enhancer factor 2A |
| chr3_+_113616317 | 0.14 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr7_-_22259845 | 0.14 |
ENST00000420196.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr11_+_12108410 | 0.14 |
ENST00000527997.1
|
RP13-631K18.5
|
RP13-631K18.5 |
| chr1_-_40042416 | 0.14 |
ENST00000372857.3
ENST00000372856.3 ENST00000531243.2 ENST00000451091.2 |
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr12_-_131200810 | 0.13 |
ENST00000536002.1
ENST00000544034.1 |
RIMBP2
RP11-662M24.2
|
RIMS binding protein 2 RP11-662M24.2 |
| chr19_-_51220176 | 0.13 |
ENST00000359082.3
ENST00000293441.1 |
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr15_+_100106155 | 0.13 |
ENST00000557785.1
ENST00000558049.1 ENST00000449277.2 |
MEF2A
|
myocyte enhancer factor 2A |
| chr7_+_80267973 | 0.13 |
ENST00000394788.3
ENST00000447544.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2C
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.2 | 1.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.2 | 4.0 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 1.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 1.0 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.5 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.4 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 0.5 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 | 1.8 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.6 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.6 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.7 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.6 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.1 | 0.3 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 0.4 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 0.2 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.1 | 0.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.9 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.7 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 1.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 0.4 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 0.3 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 5.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.4 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.2 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 0.4 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.1 | 0.9 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 1.2 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 1.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.9 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0050894 | determination of affect(GO:0050894) |
| 0.0 | 0.6 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.6 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) auditory receptor cell fate determination(GO:0042668) negative regulation of auditory receptor cell differentiation(GO:0045608) regulation of timing of neuron differentiation(GO:0060164) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.1 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.1 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.2 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.3 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.4 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 2.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.6 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 1.0 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.2 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 1.2 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.4 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 1.5 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.0 | 0.6 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.3 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.4 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.2 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0098754 | detoxification(GO:0098754) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.5 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.0 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.5 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.4 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 1.2 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0061767 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.2 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 0.4 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.1 | 1.4 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 0.3 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 1.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 4.8 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 1.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 1.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 2.5 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.6 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 0.4 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 1.0 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 0.7 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 1.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.6 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 1.0 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 2.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.6 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 1.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.6 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 0.4 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.3 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.1 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.1 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 1.0 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 1.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.3 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 1.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.6 | GO:0055103 | ligase regulator activity(GO:0055103) |
| 0.0 | 0.1 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.0 | 1.1 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0008841 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 3.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.0 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.2 | GO:0035005 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 2.7 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.5 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 7.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 2.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |