Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
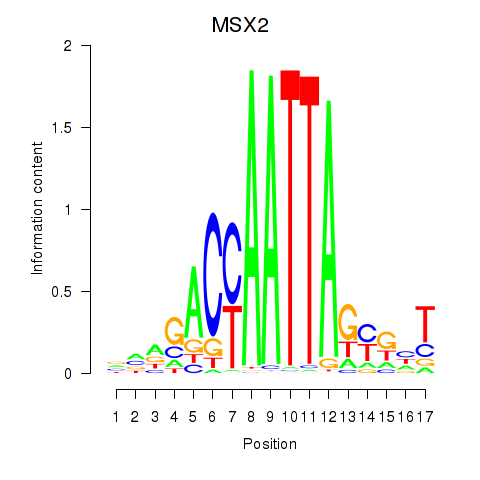
Results for MSX2
Z-value: 0.43
Transcription factors associated with MSX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MSX2
|
ENSG00000120149.7 | msh homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MSX2 | hg19_v2_chr5_+_174151536_174151610 | 0.87 | 4.9e-07 | Click! |
Activity profile of MSX2 motif
Sorted Z-values of MSX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_120400960 | 2.14 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr17_-_43045439 | 1.79 |
ENST00000253407.3
|
C1QL1
|
complement component 1, q subcomponent-like 1 |
| chr22_-_30960876 | 1.67 |
ENST00000401975.1
ENST00000428682.1 ENST00000423299.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr12_+_52695617 | 1.28 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr9_+_80912059 | 1.25 |
ENST00000347159.2
ENST00000376588.3 |
PSAT1
|
phosphoserine aminotransferase 1 |
| chr17_-_46716647 | 0.92 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr12_+_53662073 | 0.86 |
ENST00000553219.1
ENST00000257934.4 |
ESPL1
|
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr10_-_99205607 | 0.83 |
ENST00000477692.2
ENST00000485122.2 ENST00000370886.5 ENST00000370885.4 ENST00000370902.3 ENST00000370884.5 |
EXOSC1
|
exosome component 1 |
| chr12_+_53662110 | 0.73 |
ENST00000552462.1
|
ESPL1
|
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr1_-_197115818 | 0.73 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr19_+_42817450 | 0.67 |
ENST00000301204.3
|
TMEM145
|
transmembrane protein 145 |
| chr17_-_48785216 | 0.66 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr11_-_124981475 | 0.65 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr11_+_101918153 | 0.64 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr20_-_50419055 | 0.63 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr17_-_38574169 | 0.62 |
ENST00000423485.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr10_-_72142345 | 0.62 |
ENST00000373224.1
ENST00000446961.1 ENST00000358141.2 ENST00000357631.2 |
LRRC20
|
leucine rich repeat containing 20 |
| chr20_-_54967187 | 0.59 |
ENST00000422322.1
ENST00000371356.2 ENST00000451915.1 ENST00000347343.2 ENST00000395911.1 ENST00000395907.1 ENST00000441357.1 ENST00000456249.1 ENST00000420474.1 ENST00000395909.4 ENST00000395914.1 ENST00000312783.6 ENST00000395915.3 ENST00000395913.3 |
AURKA
|
aurora kinase A |
| chr15_+_80351910 | 0.59 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr12_-_120315074 | 0.55 |
ENST00000261833.7
ENST00000392521.2 |
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr14_-_95236551 | 0.53 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr15_+_41245160 | 0.53 |
ENST00000444189.2
ENST00000446533.3 |
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr17_+_42264556 | 0.47 |
ENST00000319511.6
ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr11_+_67250490 | 0.47 |
ENST00000528641.2
ENST00000279146.3 |
AIP
|
aryl hydrocarbon receptor interacting protein |
| chr5_+_162864575 | 0.47 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr8_-_12612962 | 0.43 |
ENST00000398246.3
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr12_+_9102632 | 0.43 |
ENST00000539240.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr8_-_1922789 | 0.41 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr7_-_99716952 | 0.40 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr7_-_99716940 | 0.38 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr9_-_4666421 | 0.37 |
ENST00000381895.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr9_+_34646624 | 0.36 |
ENST00000450095.2
ENST00000556278.1 |
GALT
GALT
|
galactose-1-phosphate uridylyltransferase Uncharacterized protein |
| chrX_+_77154935 | 0.36 |
ENST00000481445.1
|
COX7B
|
cytochrome c oxidase subunit VIIb |
| chr9_+_34646651 | 0.35 |
ENST00000378842.3
|
GALT
|
galactose-1-phosphate uridylyltransferase |
| chr20_-_50418947 | 0.35 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr7_+_130794878 | 0.35 |
ENST00000416992.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr17_+_12569306 | 0.34 |
ENST00000425538.1
|
MYOCD
|
myocardin |
| chr20_-_50418972 | 0.34 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr2_+_172864490 | 0.32 |
ENST00000315796.4
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr6_+_30687978 | 0.32 |
ENST00000327892.8
ENST00000435534.1 |
TUBB
|
tubulin, beta class I |
| chr7_-_99717463 | 0.31 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr5_+_140557371 | 0.30 |
ENST00000239444.2
|
PCDHB8
|
protocadherin beta 8 |
| chr1_-_151762900 | 0.30 |
ENST00000440583.2
|
TDRKH
|
tudor and KH domain containing |
| chr15_-_37393406 | 0.30 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chrX_+_70752917 | 0.30 |
ENST00000373719.3
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr12_-_57039739 | 0.29 |
ENST00000552959.1
ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr7_-_56174161 | 0.29 |
ENST00000395422.3
|
CHCHD2
|
coiled-coil-helix-coiled-coil-helix domain containing 2 |
| chr6_-_116833500 | 0.28 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr1_-_47779762 | 0.27 |
ENST00000371877.3
ENST00000360380.3 ENST00000337817.5 ENST00000447475.2 |
STIL
|
SCL/TAL1 interrupting locus |
| chr11_+_33061543 | 0.27 |
ENST00000432887.1
ENST00000528898.1 ENST00000531632.2 |
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr1_-_21625486 | 0.25 |
ENST00000481130.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr11_+_17298255 | 0.25 |
ENST00000531172.1
ENST00000533738.2 ENST00000323688.6 |
NUCB2
|
nucleobindin 2 |
| chr10_-_105110831 | 0.24 |
ENST00000337211.4
|
PCGF6
|
polycomb group ring finger 6 |
| chr10_-_105110890 | 0.24 |
ENST00000369847.3
|
PCGF6
|
polycomb group ring finger 6 |
| chr22_+_17956618 | 0.24 |
ENST00000262608.8
|
CECR2
|
cat eye syndrome chromosome region, candidate 2 |
| chr1_-_151762943 | 0.24 |
ENST00000368825.3
ENST00000368823.1 ENST00000458431.2 ENST00000368827.6 ENST00000368824.3 |
TDRKH
|
tudor and KH domain containing |
| chr1_-_190446759 | 0.24 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr19_+_42817527 | 0.24 |
ENST00000598766.1
|
TMEM145
|
transmembrane protein 145 |
| chr2_-_27603582 | 0.22 |
ENST00000323703.6
ENST00000436006.1 |
ZNF513
|
zinc finger protein 513 |
| chr19_+_11485333 | 0.22 |
ENST00000312423.2
|
SWSAP1
|
SWIM-type zinc finger 7 associated protein 1 |
| chr17_+_12569472 | 0.22 |
ENST00000343344.4
|
MYOCD
|
myocardin |
| chr7_-_2883928 | 0.22 |
ENST00000275364.3
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr1_+_81001398 | 0.21 |
ENST00000418041.1
ENST00000443104.1 |
RP5-887A10.1
|
RP5-887A10.1 |
| chr4_-_171012844 | 0.20 |
ENST00000502392.1
|
AADAT
|
aminoadipate aminotransferase |
| chr18_-_44181442 | 0.20 |
ENST00000398722.4
|
LOXHD1
|
lipoxygenase homology domains 1 |
| chr10_-_99447024 | 0.20 |
ENST00000370626.3
|
AVPI1
|
arginine vasopressin-induced 1 |
| chr4_-_138453606 | 0.20 |
ENST00000412923.2
ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18
|
protocadherin 18 |
| chr8_+_92261516 | 0.20 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr7_+_99717230 | 0.19 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr3_-_11685345 | 0.18 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr2_-_55646957 | 0.17 |
ENST00000263630.8
|
CCDC88A
|
coiled-coil domain containing 88A |
| chrX_-_21676442 | 0.17 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr2_-_55647057 | 0.17 |
ENST00000436346.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr1_-_158173659 | 0.17 |
ENST00000415019.1
|
RP11-404O13.5
|
RP11-404O13.5 |
| chr19_-_19249255 | 0.16 |
ENST00000587583.2
ENST00000450333.2 ENST00000587096.1 ENST00000162044.9 ENST00000592369.1 ENST00000587915.1 |
TMEM161A
|
transmembrane protein 161A |
| chr11_-_121986923 | 0.15 |
ENST00000560104.1
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr6_+_25652432 | 0.14 |
ENST00000377961.2
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr11_+_17298522 | 0.14 |
ENST00000529313.1
|
NUCB2
|
nucleobindin 2 |
| chrX_+_70752945 | 0.14 |
ENST00000373701.3
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr11_+_17298543 | 0.14 |
ENST00000533926.1
|
NUCB2
|
nucleobindin 2 |
| chr4_-_19458597 | 0.13 |
ENST00000505347.1
|
RP11-3J1.1
|
RP11-3J1.1 |
| chr3_+_186353756 | 0.13 |
ENST00000431018.1
ENST00000450521.1 ENST00000539949.1 |
FETUB
|
fetuin B |
| chr1_-_232651312 | 0.13 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr11_+_17298297 | 0.13 |
ENST00000529010.1
|
NUCB2
|
nucleobindin 2 |
| chr17_-_38956205 | 0.13 |
ENST00000306658.7
|
KRT28
|
keratin 28 |
| chr2_+_90248739 | 0.12 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr5_-_132200477 | 0.12 |
ENST00000296875.2
|
GDF9
|
growth differentiation factor 9 |
| chr2_-_98280383 | 0.12 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr12_+_28410128 | 0.11 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr7_+_48211048 | 0.11 |
ENST00000435803.1
|
ABCA13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr8_+_9413410 | 0.11 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr6_+_25652501 | 0.10 |
ENST00000334979.6
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr5_-_148758839 | 0.10 |
ENST00000261796.3
|
IL17B
|
interleukin 17B |
| chr2_-_73460334 | 0.10 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1 |
| chrX_-_15332665 | 0.09 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr13_-_103389159 | 0.09 |
ENST00000322527.2
|
CCDC168
|
coiled-coil domain containing 168 |
| chr10_-_56561022 | 0.08 |
ENST00000373965.2
ENST00000414778.1 ENST00000395438.1 ENST00000409834.1 ENST00000395445.1 ENST00000395446.1 ENST00000395442.1 ENST00000395440.1 ENST00000395432.2 ENST00000361849.3 ENST00000395433.1 ENST00000320301.6 ENST00000395430.1 ENST00000437009.1 |
PCDH15
|
protocadherin-related 15 |
| chr15_+_96904487 | 0.08 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr19_-_10121144 | 0.08 |
ENST00000264828.3
|
COL5A3
|
collagen, type V, alpha 3 |
| chr1_+_202317855 | 0.08 |
ENST00000356764.2
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr1_-_151431909 | 0.08 |
ENST00000361398.3
ENST00000271715.2 |
POGZ
|
pogo transposable element with ZNF domain |
| chr3_-_112127981 | 0.08 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr6_+_34204642 | 0.07 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr14_-_51027838 | 0.07 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr1_+_244515930 | 0.07 |
ENST00000366537.1
ENST00000308105.4 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr17_-_9929581 | 0.07 |
ENST00000437099.2
ENST00000396115.2 |
GAS7
|
growth arrest-specific 7 |
| chr1_+_175036966 | 0.05 |
ENST00000239462.4
|
TNN
|
tenascin N |
| chr7_+_130794846 | 0.05 |
ENST00000421797.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr12_+_54718904 | 0.05 |
ENST00000262061.2
ENST00000549043.1 ENST00000552218.1 ENST00000553231.1 ENST00000552362.1 ENST00000455864.2 ENST00000416254.2 ENST00000549116.1 ENST00000551779.1 |
COPZ1
|
coatomer protein complex, subunit zeta 1 |
| chr21_-_22175341 | 0.05 |
ENST00000416768.1
ENST00000452561.1 ENST00000419299.1 ENST00000437238.1 |
LINC00320
|
long intergenic non-protein coding RNA 320 |
| chr18_+_63273310 | 0.05 |
ENST00000579862.1
|
RP11-775G23.1
|
RP11-775G23.1 |
| chr17_-_80250663 | 0.05 |
ENST00000581489.1
|
RP13-516M14.4
|
RP13-516M14.4 |
| chr7_-_99716914 | 0.04 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr19_-_46088068 | 0.04 |
ENST00000263275.4
ENST00000323060.3 |
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chrX_-_69269788 | 0.04 |
ENST00000276101.3
|
AWAT2
|
acyl-CoA wax alcohol acyltransferase 2 |
| chr2_-_8715616 | 0.04 |
ENST00000418358.1
|
AC011747.3
|
AC011747.3 |
| chr11_-_40315640 | 0.04 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr10_-_15762124 | 0.04 |
ENST00000378076.3
|
ITGA8
|
integrin, alpha 8 |
| chr4_-_88449771 | 0.03 |
ENST00000535835.1
|
SPARCL1
|
SPARC-like 1 (hevin) |
| chr15_-_51535208 | 0.03 |
ENST00000405913.3
ENST00000559878.1 |
CYP19A1
|
cytochrome P450, family 19, subfamily A, polypeptide 1 |
| chr3_+_129247479 | 0.02 |
ENST00000296271.3
|
RHO
|
rhodopsin |
| chr6_-_139613269 | 0.02 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chr11_-_71823266 | 0.02 |
ENST00000538919.1
ENST00000539395.1 ENST00000542531.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr11_-_102323740 | 0.01 |
ENST00000398136.2
|
TMEM123
|
transmembrane protein 123 |
| chr4_-_46996424 | 0.01 |
ENST00000264318.3
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4 |
| chr16_-_29934558 | 0.01 |
ENST00000568995.1
ENST00000566413.1 |
KCTD13
|
potassium channel tetramerization domain containing 13 |
| chr2_-_200335983 | 0.01 |
ENST00000457245.1
|
SATB2
|
SATB homeobox 2 |
| chr14_+_65381079 | 0.01 |
ENST00000549115.1
ENST00000607599.1 ENST00000548752.2 ENST00000359118.2 ENST00000552002.2 ENST00000551947.1 ENST00000551093.1 ENST00000542227.1 ENST00000447296.2 ENST00000549987.1 |
CHURC1
FNTB
CHURC1-FNTB
|
churchill domain containing 1 farnesyltransferase, CAAX box, beta CHURC1-FNTB readthrough |
| chr2_+_187371440 | 0.01 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr7_+_100209725 | 0.00 |
ENST00000223054.4
|
MOSPD3
|
motile sperm domain containing 3 |
| chr6_-_116866773 | 0.00 |
ENST00000368602.3
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of MSX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.4 | 1.7 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.2 | 0.7 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.2 | 2.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 0.7 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 0.2 | 1.3 | GO:0042816 | vitamin B6 metabolic process(GO:0042816) |
| 0.2 | 0.6 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.1 | 0.6 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 1.7 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.9 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.4 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.3 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 1.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.6 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.5 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.3 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.2 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) L-kynurenine metabolic process(GO:0097052) |
| 0.0 | 0.3 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.3 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.5 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.3 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.6 | GO:0072663 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.0 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 1.1 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.2 | 0.6 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 0.6 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.5 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 0.7 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.2 | GO:0097196 | Shu complex(GO:0097196) |
| 0.1 | 0.8 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 1.1 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.3 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 1.7 | GO:0072686 | mitotic spindle(GO:0072686) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 1.7 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.5 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.4 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 1.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.2 | GO:0047536 | 2-aminoadipate transaminase activity(GO:0047536) |
| 0.1 | 0.6 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 2.1 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 1.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.7 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.3 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.8 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.6 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |