Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
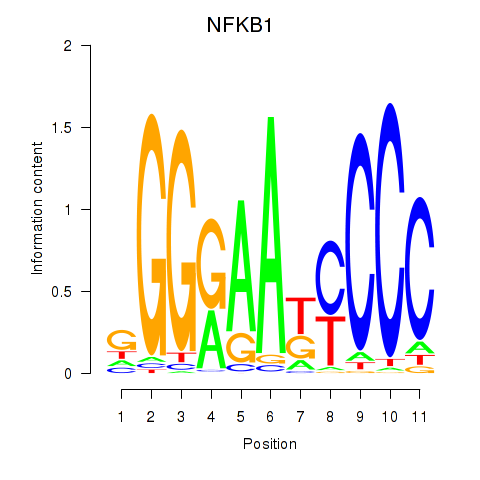
Results for NFKB1
Z-value: 1.44
Transcription factors associated with NFKB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFKB1
|
ENSG00000109320.7 | nuclear factor kappa B subunit 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFKB1 | hg19_v2_chr4_+_103422499_103422632 | -0.74 | 2.0e-04 | Click! |
Activity profile of NFKB1 motif
Sorted Z-values of NFKB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_52445191 | 12.87 |
ENST00000243050.1
ENST00000394825.1 ENST00000550763.1 ENST00000394824.2 ENST00000548232.1 ENST00000562373.1 |
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr4_-_74904398 | 8.36 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chr4_-_74964904 | 7.54 |
ENST00000508487.2
|
CXCL2
|
chemokine (C-X-C motif) ligand 2 |
| chr2_+_228678550 | 5.94 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr19_-_6591113 | 5.24 |
ENST00000423145.3
ENST00000245903.3 |
CD70
|
CD70 molecule |
| chr4_+_74735102 | 5.04 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr4_-_74864386 | 4.51 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr12_+_52430894 | 4.26 |
ENST00000546842.1
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr1_-_186649543 | 3.90 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr19_+_45504688 | 3.83 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr16_+_29823427 | 3.61 |
ENST00000358758.7
ENST00000567659.1 ENST00000572820.1 |
PRRT2
|
proline-rich transmembrane protein 2 |
| chr1_+_110453203 | 3.57 |
ENST00000357302.4
ENST00000344188.5 ENST00000329608.6 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr12_-_49259643 | 3.56 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr6_-_31550192 | 3.46 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr1_+_6845384 | 3.45 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr10_+_30723105 | 3.09 |
ENST00000375322.2
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr10_+_104154229 | 3.00 |
ENST00000428099.1
ENST00000369966.3 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr6_-_29527702 | 2.97 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr12_+_7052974 | 2.92 |
ENST00000544681.1
ENST00000537087.1 |
C12orf57
|
chromosome 12 open reading frame 57 |
| chr19_-_46145696 | 2.89 |
ENST00000588172.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr1_+_186649754 | 2.84 |
ENST00000608917.1
|
RP5-973M2.2
|
RP5-973M2.2 |
| chr14_-_24616426 | 2.84 |
ENST00000216802.5
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr1_+_37940153 | 2.82 |
ENST00000373087.6
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr1_+_201979743 | 2.76 |
ENST00000446188.1
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr10_+_30723045 | 2.66 |
ENST00000542547.1
ENST00000415139.1 |
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr14_+_24521202 | 2.65 |
ENST00000334420.7
ENST00000342740.5 |
LRRC16B
|
leucine rich repeat containing 16B |
| chr19_-_41222775 | 2.57 |
ENST00000324464.3
ENST00000450541.1 ENST00000594720.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr14_-_103589246 | 2.54 |
ENST00000558224.1
ENST00000560742.1 |
LINC00677
|
long intergenic non-protein coding RNA 677 |
| chr19_+_55795493 | 2.53 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr10_+_30722866 | 2.52 |
ENST00000263056.1
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr12_+_7053172 | 2.50 |
ENST00000229281.5
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr6_-_30080876 | 2.48 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr1_-_41950342 | 2.47 |
ENST00000372587.4
|
EDN2
|
endothelin 2 |
| chr6_+_32821924 | 2.44 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr19_+_42364313 | 2.44 |
ENST00000601492.1
ENST00000600467.1 ENST00000221975.2 |
RPS19
|
ribosomal protein S19 |
| chr19_+_42363917 | 2.44 |
ENST00000598742.1
|
RPS19
|
ribosomal protein S19 |
| chr6_-_30080863 | 2.42 |
ENST00000540829.1
|
TRIM31
|
tripartite motif containing 31 |
| chr19_-_38747172 | 2.40 |
ENST00000347262.4
ENST00000591585.1 ENST00000301242.4 |
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr1_+_201979645 | 2.36 |
ENST00000367284.5
ENST00000367283.3 |
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr2_-_157189180 | 2.27 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr10_-_7708918 | 2.25 |
ENST00000256861.6
ENST00000397146.2 ENST00000446830.2 ENST00000397145.2 |
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr1_+_110453514 | 2.19 |
ENST00000369802.3
ENST00000420111.2 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr12_-_58131931 | 2.18 |
ENST00000547588.1
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr17_+_4854375 | 2.16 |
ENST00000521811.1
ENST00000519602.1 ENST00000323997.6 ENST00000522249.1 ENST00000519584.1 |
ENO3
|
enolase 3 (beta, muscle) |
| chr19_-_38746979 | 2.14 |
ENST00000591291.1
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr6_+_138188351 | 2.12 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr11_+_560956 | 2.11 |
ENST00000397582.3
ENST00000344375.4 ENST00000397583.3 |
RASSF7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr11_+_72525353 | 2.09 |
ENST00000321297.5
ENST00000534905.1 ENST00000540567.1 |
ATG16L2
|
autophagy related 16-like 2 (S. cerevisiae) |
| chr19_-_46272462 | 2.08 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr12_+_7053228 | 2.07 |
ENST00000540506.2
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr6_+_29691056 | 2.05 |
ENST00000414333.1
ENST00000334668.4 ENST00000259951.7 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr19_+_42364460 | 2.03 |
ENST00000593863.1
|
RPS19
|
ribosomal protein S19 |
| chr19_+_35485682 | 2.01 |
ENST00000599564.1
|
GRAMD1A
|
GRAM domain containing 1A |
| chr19_+_2476116 | 2.00 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr14_-_35873856 | 1.98 |
ENST00000553342.1
ENST00000216797.5 ENST00000557140.1 |
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| chr19_-_4182530 | 1.98 |
ENST00000601571.1
ENST00000601488.1 ENST00000305232.6 ENST00000381935.3 ENST00000337491.2 |
SIRT6
|
sirtuin 6 |
| chr12_-_62997214 | 1.96 |
ENST00000408887.2
|
C12orf61
|
chromosome 12 open reading frame 61 |
| chr7_+_69064566 | 1.96 |
ENST00000403018.2
|
AUTS2
|
autism susceptibility candidate 2 |
| chr1_+_6845497 | 1.93 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr19_-_47734448 | 1.91 |
ENST00000439096.2
|
BBC3
|
BCL2 binding component 3 |
| chr11_-_3862206 | 1.87 |
ENST00000351018.4
|
RHOG
|
ras homolog family member G |
| chr19_+_48898132 | 1.85 |
ENST00000263269.3
|
GRIN2D
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D |
| chr7_-_100881109 | 1.82 |
ENST00000308344.5
|
CLDN15
|
claudin 15 |
| chr11_-_66313699 | 1.82 |
ENST00000526986.1
ENST00000310442.3 |
ZDHHC24
|
zinc finger, DHHC-type containing 24 |
| chr11_+_102188272 | 1.81 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr17_+_7123207 | 1.80 |
ENST00000584103.1
ENST00000579886.2 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr19_+_40697514 | 1.76 |
ENST00000253055.3
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr9_-_136344197 | 1.75 |
ENST00000414172.1
ENST00000371897.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr19_-_46272106 | 1.72 |
ENST00000560168.1
|
SIX5
|
SIX homeobox 5 |
| chr12_+_56732658 | 1.69 |
ENST00000228534.4
|
IL23A
|
interleukin 23, alpha subunit p19 |
| chr6_-_33160231 | 1.69 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr2_-_177502659 | 1.68 |
ENST00000295549.4
|
AC017048.3
|
long intergenic non-protein coding RNA 1116 |
| chr16_+_29823552 | 1.63 |
ENST00000300797.6
|
PRRT2
|
proline-rich transmembrane protein 2 |
| chr17_+_47074758 | 1.63 |
ENST00000290341.3
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr6_+_29691198 | 1.62 |
ENST00000440587.2
ENST00000434407.2 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr15_+_76030311 | 1.59 |
ENST00000543887.1
|
AC019294.1
|
AC019294.1 |
| chr19_+_45251804 | 1.57 |
ENST00000164227.5
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr11_+_66059339 | 1.55 |
ENST00000327259.4
|
TMEM151A
|
transmembrane protein 151A |
| chr10_+_89419370 | 1.52 |
ENST00000361175.4
ENST00000456849.1 |
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr17_+_25799008 | 1.51 |
ENST00000583370.1
ENST00000398988.3 ENST00000268763.6 |
KSR1
|
kinase suppressor of ras 1 |
| chr19_-_4182497 | 1.50 |
ENST00000597896.1
|
SIRT6
|
sirtuin 6 |
| chr20_+_44746885 | 1.47 |
ENST00000372285.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr12_+_52431016 | 1.46 |
ENST00000553200.1
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr1_+_6845578 | 1.45 |
ENST00000467404.2
ENST00000439411.2 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr1_+_38259540 | 1.42 |
ENST00000397631.3
|
MANEAL
|
mannosidase, endo-alpha-like |
| chr5_-_137090028 | 1.41 |
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr17_-_61777090 | 1.40 |
ENST00000578061.1
|
LIMD2
|
LIM domain containing 2 |
| chr14_+_24439148 | 1.39 |
ENST00000543805.1
ENST00000534993.1 |
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chr20_-_2451395 | 1.35 |
ENST00000339610.6
ENST00000381342.2 ENST00000438552.2 |
SNRPB
|
small nuclear ribonucleoprotein polypeptides B and B1 |
| chr19_+_41860047 | 1.29 |
ENST00000604123.1
|
TMEM91
|
transmembrane protein 91 |
| chr2_+_97202480 | 1.29 |
ENST00000357485.3
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr9_+_134103496 | 1.28 |
ENST00000498010.1
ENST00000476004.1 ENST00000528406.1 |
NUP214
|
nucleoporin 214kDa |
| chr7_-_93520191 | 1.27 |
ENST00000545378.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr3_+_49058444 | 1.26 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr17_+_7590734 | 1.26 |
ENST00000457584.2
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr2_+_38893047 | 1.21 |
ENST00000272252.5
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr12_+_6875519 | 1.21 |
ENST00000389462.4
ENST00000540874.1 ENST00000309083.6 |
PTMS
|
parathymosin |
| chr3_+_52280173 | 1.20 |
ENST00000296487.4
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr19_-_17958832 | 1.19 |
ENST00000458235.1
|
JAK3
|
Janus kinase 3 |
| chr6_+_138188551 | 1.19 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr1_-_205904950 | 1.19 |
ENST00000340781.4
|
SLC26A9
|
solute carrier family 26 (anion exchanger), member 9 |
| chr14_-_24615805 | 1.18 |
ENST00000560410.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr20_+_44746939 | 1.18 |
ENST00000372276.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr1_+_38259459 | 1.18 |
ENST00000373045.6
|
MANEAL
|
mannosidase, endo-alpha-like |
| chr8_+_145582217 | 1.18 |
ENST00000530047.1
ENST00000527078.1 |
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chr11_+_63997750 | 1.17 |
ENST00000321685.3
|
DNAJC4
|
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr7_-_100881041 | 1.17 |
ENST00000412417.1
ENST00000414035.1 |
CLDN15
|
claudin 15 |
| chr19_-_46234119 | 1.16 |
ENST00000317683.3
|
FBXO46
|
F-box protein 46 |
| chr10_+_102756800 | 1.16 |
ENST00000370223.3
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr6_-_32122106 | 1.15 |
ENST00000428778.1
|
PRRT1
|
proline-rich transmembrane protein 1 |
| chr11_-_6426635 | 1.15 |
ENST00000608645.1
ENST00000608394.1 ENST00000529519.1 |
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr2_+_97203082 | 1.14 |
ENST00000454558.2
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr17_+_48638371 | 1.12 |
ENST00000360761.4
ENST00000352832.5 ENST00000354983.4 |
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr17_+_30594823 | 1.09 |
ENST00000536287.1
|
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr8_-_145582118 | 1.09 |
ENST00000455319.2
ENST00000331890.5 |
FBXL6
|
F-box and leucine-rich repeat protein 6 |
| chr2_+_38893208 | 1.07 |
ENST00000410063.1
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr11_-_64570706 | 1.07 |
ENST00000294066.2
ENST00000377350.3 |
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr16_+_57126428 | 1.07 |
ENST00000290776.8
|
CPNE2
|
copine II |
| chrX_-_100129128 | 1.06 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr3_+_52280220 | 1.05 |
ENST00000409502.3
ENST00000323588.4 |
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr17_+_47075023 | 1.02 |
ENST00000431824.2
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr1_+_169077172 | 1.02 |
ENST00000499679.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr17_-_4852332 | 1.01 |
ENST00000572383.1
|
PFN1
|
profilin 1 |
| chr1_-_8000872 | 1.01 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr11_+_119056178 | 1.01 |
ENST00000525131.1
ENST00000531114.1 ENST00000355547.5 ENST00000322712.4 |
PDZD3
|
PDZ domain containing 3 |
| chr19_-_33555780 | 1.01 |
ENST00000254260.3
ENST00000400226.4 |
RHPN2
|
rhophilin, Rho GTPase binding protein 2 |
| chr19_+_18492973 | 1.00 |
ENST00000595973.2
|
GDF15
|
growth differentiation factor 15 |
| chr19_-_40324255 | 1.00 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chrX_-_100129320 | 0.99 |
ENST00000372966.3
|
NOX1
|
NADPH oxidase 1 |
| chr17_+_2264983 | 0.98 |
ENST00000574650.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chr11_-_75062730 | 0.97 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr8_+_145582231 | 0.96 |
ENST00000526338.1
ENST00000402965.1 ENST00000534725.1 ENST00000532887.1 ENST00000329994.2 |
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chr1_+_110453462 | 0.95 |
ENST00000488198.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr7_-_100808394 | 0.95 |
ENST00000445482.2
|
VGF
|
VGF nerve growth factor inducible |
| chr19_-_5785630 | 0.95 |
ENST00000586012.1
ENST00000590343.1 |
CTB-54O9.9
DUS3L
|
Uncharacterized protein dihydrouridine synthase 3-like (S. cerevisiae) |
| chr16_+_57126482 | 0.94 |
ENST00000537605.1
ENST00000535318.2 |
CPNE2
|
copine II |
| chr7_-_155601766 | 0.93 |
ENST00000430104.1
|
SHH
|
sonic hedgehog |
| chr1_-_21978312 | 0.93 |
ENST00000359708.4
ENST00000290101.4 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr11_-_44331679 | 0.93 |
ENST00000329255.3
|
ALX4
|
ALX homeobox 4 |
| chr11_-_46142948 | 0.93 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chr17_+_40440481 | 0.92 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr17_+_4634705 | 0.92 |
ENST00000575284.1
ENST00000573708.1 ENST00000293777.5 |
MED11
|
mediator complex subunit 11 |
| chr2_-_217560248 | 0.92 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr7_-_93520259 | 0.90 |
ENST00000222543.5
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr5_+_159895275 | 0.90 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr2_-_163175133 | 0.88 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr14_-_64970494 | 0.88 |
ENST00000608382.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr17_-_7154984 | 0.87 |
ENST00000574322.1
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr19_+_1067271 | 0.87 |
ENST00000536472.1
ENST00000590214.1 |
HMHA1
|
histocompatibility (minor) HA-1 |
| chr17_-_7123021 | 0.87 |
ENST00000399510.2
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr15_-_27018884 | 0.86 |
ENST00000299267.4
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr11_+_118754475 | 0.86 |
ENST00000292174.4
|
CXCR5
|
chemokine (C-X-C motif) receptor 5 |
| chr17_-_17740287 | 0.86 |
ENST00000355815.4
ENST00000261646.5 |
SREBF1
|
sterol regulatory element binding transcription factor 1 |
| chr16_+_67282853 | 0.86 |
ENST00000299798.11
|
SLC9A5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr19_+_39390320 | 0.85 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr1_+_45274154 | 0.85 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr19_+_39390587 | 0.84 |
ENST00000572515.1
ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr6_+_135502466 | 0.84 |
ENST00000367814.4
|
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr11_+_102188224 | 0.83 |
ENST00000263464.3
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr2_+_163175394 | 0.82 |
ENST00000446271.1
ENST00000429691.2 |
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr3_-_156878482 | 0.81 |
ENST00000295925.4
|
CCNL1
|
cyclin L1 |
| chr17_-_39968406 | 0.80 |
ENST00000393928.1
|
LEPREL4
|
leprecan-like 4 |
| chr5_-_127418573 | 0.80 |
ENST00000508353.1
ENST00000508878.1 ENST00000501652.1 ENST00000514409.1 |
CTC-228N24.3
|
CTC-228N24.3 |
| chr4_+_185395947 | 0.80 |
ENST00000605834.1
|
RP11-326I11.3
|
RP11-326I11.3 |
| chr3_-_156878540 | 0.79 |
ENST00000461804.1
|
CCNL1
|
cyclin L1 |
| chr5_+_170846640 | 0.79 |
ENST00000274625.5
|
FGF18
|
fibroblast growth factor 18 |
| chr1_+_56880606 | 0.79 |
ENST00000451914.1
|
RP4-710M16.2
|
RP4-710M16.2 |
| chr11_+_65292538 | 0.77 |
ENST00000270176.5
ENST00000525364.1 ENST00000420247.2 ENST00000533862.1 ENST00000279270.6 ENST00000524944.1 |
SCYL1
|
SCY1-like 1 (S. cerevisiae) |
| chr20_-_62462566 | 0.77 |
ENST00000245663.4
ENST00000302995.2 |
ZBTB46
|
zinc finger and BTB domain containing 46 |
| chr17_+_7123125 | 0.77 |
ENST00000356839.5
ENST00000583312.1 ENST00000350303.5 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr1_+_185703513 | 0.77 |
ENST00000271588.4
ENST00000367492.2 |
HMCN1
|
hemicentin 1 |
| chr19_-_39330818 | 0.76 |
ENST00000594769.1
ENST00000602021.1 |
AC104534.3
|
Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial |
| chr16_-_4303767 | 0.75 |
ENST00000573268.1
ENST00000573042.1 |
RP11-95P2.1
|
RP11-95P2.1 |
| chr17_-_1420006 | 0.75 |
ENST00000320345.6
ENST00000406424.4 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr8_+_72755796 | 0.75 |
ENST00000524152.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548 |
| chr15_-_89438742 | 0.74 |
ENST00000562281.1
ENST00000562889.1 ENST00000359595.3 |
HAPLN3
|
hyaluronan and proteoglycan link protein 3 |
| chr6_-_32821599 | 0.73 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr10_+_99609996 | 0.73 |
ENST00000370602.1
|
GOLGA7B
|
golgin A7 family, member B |
| chr1_+_159796534 | 0.73 |
ENST00000289707.5
|
SLAMF8
|
SLAM family member 8 |
| chr12_+_49761224 | 0.73 |
ENST00000553127.1
ENST00000321898.6 |
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr19_+_1067144 | 0.72 |
ENST00000313093.2
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chr10_+_12391481 | 0.71 |
ENST00000378847.3
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr4_-_122085469 | 0.70 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr6_-_31138439 | 0.70 |
ENST00000259915.8
|
POU5F1
|
POU class 5 homeobox 1 |
| chr4_+_142557717 | 0.69 |
ENST00000320650.4
ENST00000296545.7 |
IL15
|
interleukin 15 |
| chr17_+_79373540 | 0.69 |
ENST00000307745.7
|
RP11-1055B8.7
|
BAH and coiled-coil domain-containing protein 1 |
| chr9_-_140115775 | 0.69 |
ENST00000391553.1
ENST00000392827.1 |
RNF208
|
ring finger protein 208 |
| chr5_-_150460914 | 0.68 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr19_+_39759154 | 0.67 |
ENST00000331982.5
|
IFNL2
|
interferon, lambda 2 |
| chr6_+_30297306 | 0.66 |
ENST00000420746.1
ENST00000513556.1 |
TRIM39
TRIM39-RPP21
|
tripartite motif containing 39 TRIM39-RPP21 readthrough |
| chr1_-_24469602 | 0.66 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1 |
| chr5_-_127418755 | 0.66 |
ENST00000501702.2
ENST00000501173.2 ENST00000514573.1 ENST00000499346.2 ENST00000606251.1 |
CTC-228N24.3
|
CTC-228N24.3 |
| chr19_-_39735646 | 0.66 |
ENST00000413851.2
|
IFNL3
|
interferon, lambda 3 |
| chr12_-_132905789 | 0.65 |
ENST00000328957.8
|
GALNT9
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 9 (GalNAc-T9) |
| chr17_-_1420182 | 0.64 |
ENST00000421807.2
|
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr5_-_67730240 | 0.63 |
ENST00000507733.1
|
CTC-537E7.3
|
CTC-537E7.3 |
| chrX_-_21776281 | 0.63 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr2_-_100721178 | 0.63 |
ENST00000409236.2
|
AFF3
|
AF4/FMR2 family, member 3 |
| chr19_+_1067492 | 0.63 |
ENST00000586866.1
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chr1_+_221054584 | 0.62 |
ENST00000549319.1
|
HLX
|
H2.0-like homeobox |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFKB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 1.7 | 6.9 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 1.6 | 20.9 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 1.5 | 7.5 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 1.4 | 7.1 | GO:1903971 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 1.3 | 3.9 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 1.2 | 3.5 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) histone H3-K9 deacetylation(GO:1990619) |
| 1.1 | 4.6 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 1.0 | 3.1 | GO:2000349 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.9 | 3.8 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.9 | 2.8 | GO:2000625 | regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.7 | 2.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.6 | 2.5 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.6 | 2.3 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.5 | 1.6 | GO:0045210 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 0.5 | 14.0 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.5 | 2.6 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.5 | 2.0 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.5 | 1.5 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.4 | 3.5 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.4 | 2.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.4 | 1.3 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.4 | 3.9 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.4 | 1.1 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.4 | 1.5 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.4 | 6.0 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.3 | 2.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.3 | 1.7 | GO:0060023 | soft palate development(GO:0060023) |
| 0.3 | 1.3 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.3 | 1.9 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.3 | 0.9 | GO:0060738 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of immature T cell proliferation(GO:0033091) positive regulation of immature T cell proliferation in thymus(GO:0033092) right lung development(GO:0060458) primary prostatic bud elongation(GO:0060516) epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.3 | 0.9 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.3 | 0.8 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.3 | 3.0 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.3 | 2.6 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.3 | 2.9 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.2 | 5.2 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.2 | 1.5 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.2 | 1.0 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.2 | 0.7 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.2 | 0.9 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.2 | 0.4 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.2 | 0.6 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.2 | 1.0 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.2 | 1.8 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 0.7 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.2 | 1.2 | GO:1903288 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.2 | 3.6 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.2 | 1.9 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 1.0 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.2 | 5.2 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.2 | 1.0 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.2 | 2.6 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.2 | 0.6 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.2 | 0.5 | GO:0090330 | regulation of platelet aggregation(GO:0090330) negative regulation of platelet aggregation(GO:0090331) |
| 0.2 | 0.8 | GO:0031296 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.1 | 0.4 | GO:1903093 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 | 1.6 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.5 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 9.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.8 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 1.0 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.8 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.1 | 0.6 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 1.0 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.7 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 | 7.9 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 0.9 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 0.3 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 | 0.6 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.6 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.2 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.1 | 1.0 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.2 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.6 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.1 | 2.8 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.4 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.1 | 1.3 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 1.7 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.2 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 0.3 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 1.4 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 1.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.2 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.5 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.1 | 0.2 | GO:0031133 | regulation of axon diameter(GO:0031133) regulation of adenosine receptor signaling pathway(GO:0060167) positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.1 | 0.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 2.2 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 1.9 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.1 | 0.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.9 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.4 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.8 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0090291 | negative regulation of osteoclast proliferation(GO:0090291) |
| 0.0 | 0.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 2.2 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.5 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 1.8 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 1.0 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 4.1 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.6 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 1.0 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.9 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 1.6 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 4.2 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 2.4 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 1.8 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.6 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.6 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.4 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.4 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.7 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 0.9 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.3 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 1.3 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.7 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 1.9 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.6 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.8 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 0.0 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.3 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.7 | GO:0071219 | cellular response to molecule of bacterial origin(GO:0071219) |
| 0.0 | 0.1 | GO:1901660 | calcium ion export(GO:1901660) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.0 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.3 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.7 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.4 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 1.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.9 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.3 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.9 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 1.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.9 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 2.0 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.6 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 1.0 | 7.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.9 | 3.5 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.7 | 4.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.4 | 2.8 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.4 | 3.9 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.3 | 2.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.3 | 2.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 1.3 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.2 | 2.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 1.3 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 0.9 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 1.9 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 1.3 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 1.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 2.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 1.7 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.6 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.7 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.6 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 6.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.6 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 1.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 0.4 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.1 | 1.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 1.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 4.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.7 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 2.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 3.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.6 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 2.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 3.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 3.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 1.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 11.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 4.5 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 24.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.8 | GO:0032587 | ruffle membrane(GO:0032587) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 1.8 | 26.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 1.2 | 3.5 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.9 | 7.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.8 | 3.9 | GO:0050473 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.6 | 4.4 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.5 | 1.5 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.5 | 2.5 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.5 | 1.5 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.4 | 4.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.4 | 1.8 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.4 | 2.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.4 | 1.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.3 | 2.0 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.3 | 2.6 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.3 | 1.0 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.2 | 2.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.2 | 1.9 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 3.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 1.3 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.2 | 7.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 0.6 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.2 | 6.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.2 | 21.4 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.2 | 9.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 1.0 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.2 | 1.7 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.2 | 0.6 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 2.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 1.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 2.8 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.7 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.1 | 0.9 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 2.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 1.0 | GO:0030249 | cyclase regulator activity(GO:0010851) guanylate cyclase regulator activity(GO:0030249) |
| 0.1 | 2.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.9 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.4 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.1 | 3.7 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 1.8 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 2.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.2 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.1 | 0.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 1.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.4 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.1 | 1.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.9 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 2.7 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 1.0 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 2.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 4.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.9 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.8 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.9 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 1.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 2.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.8 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 1.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 3.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 1.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.7 | GO:0001054 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 4.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 1.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 8.9 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 3.5 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 2.6 | GO:0016798 | hydrolase activity, acting on glycosyl bonds(GO:0016798) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.9 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.0 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 1.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.3 | 20.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.3 | 8.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 7.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 9.9 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 8.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 3.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 6.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 2.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 2.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 24.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.7 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.5 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.2 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 33.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 3.9 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.3 | 8.3 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.2 | 11.8 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.2 | 7.5 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.2 | 3.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 2.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 6.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 1.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 1.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 5.0 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 0.5 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 1.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 2.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.9 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 1.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 2.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 2.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.9 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 3.7 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 2.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 2.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |