Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
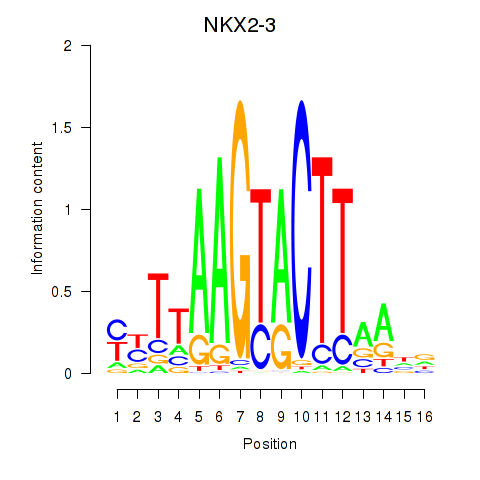
Results for NKX2-3
Z-value: 0.59
Transcription factors associated with NKX2-3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-3
|
ENSG00000119919.9 | NK2 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-3 | hg19_v2_chr10_+_101292684_101292706 | -0.17 | 4.6e-01 | Click! |
Activity profile of NKX2-3 motif
Sorted Z-values of NKX2-3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_150404904 | 2.15 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr4_-_90758227 | 0.88 |
ENST00000506691.1
ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr11_+_35211511 | 0.87 |
ENST00000524922.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr12_-_123201337 | 0.84 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr1_+_35220613 | 0.78 |
ENST00000338513.1
|
GJB5
|
gap junction protein, beta 5, 31.1kDa |
| chr5_+_179105615 | 0.76 |
ENST00000514383.1
|
CANX
|
calnexin |
| chr17_+_66521936 | 0.76 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr12_-_123187890 | 0.75 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr18_-_33702078 | 0.74 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr18_-_74839891 | 0.72 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr16_-_69166031 | 0.70 |
ENST00000398235.2
ENST00000520529.1 |
CHTF8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae) |
| chr7_-_2883650 | 0.69 |
ENST00000544127.1
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr18_+_72166564 | 0.69 |
ENST00000583216.1
ENST00000581912.1 ENST00000582589.1 |
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr7_-_80548493 | 0.69 |
ENST00000536800.1
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr19_-_51504852 | 0.67 |
ENST00000391806.2
ENST00000347619.4 ENST00000291726.7 ENST00000320838.5 |
KLK8
|
kallikrein-related peptidase 8 |
| chr17_+_26662679 | 0.67 |
ENST00000578158.1
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr9_+_131447342 | 0.67 |
ENST00000409104.3
|
SET
|
SET nuclear oncogene |
| chr19_+_7445850 | 0.66 |
ENST00000593531.1
|
CTD-2207O23.3
|
Rho guanine nucleotide exchange factor 18 |
| chr10_-_32345305 | 0.64 |
ENST00000302418.4
|
KIF5B
|
kinesin family member 5B |
| chr3_-_69062790 | 0.63 |
ENST00000540955.1
ENST00000456376.1 |
EOGT
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr7_-_80548667 | 0.63 |
ENST00000265361.3
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr3_+_113775594 | 0.62 |
ENST00000479882.1
ENST00000493014.1 |
QTRTD1
|
queuine tRNA-ribosyltransferase domain containing 1 |
| chr11_+_35211429 | 0.62 |
ENST00000525688.1
ENST00000278385.6 ENST00000533222.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr1_+_230193521 | 0.62 |
ENST00000543760.1
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr2_+_64073187 | 0.62 |
ENST00000491621.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr5_-_159846066 | 0.61 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chrX_+_114795489 | 0.61 |
ENST00000355899.3
ENST00000537301.1 ENST00000289290.3 |
PLS3
|
plastin 3 |
| chr14_-_69262947 | 0.59 |
ENST00000557086.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr6_-_26216872 | 0.59 |
ENST00000244601.3
|
HIST1H2BG
|
histone cluster 1, H2bg |
| chr4_-_85654615 | 0.58 |
ENST00000514711.1
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr10_+_101419187 | 0.57 |
ENST00000370489.4
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr2_-_178128528 | 0.57 |
ENST00000397063.4
ENST00000421929.1 |
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr4_+_48343339 | 0.57 |
ENST00000264313.6
|
SLAIN2
|
SLAIN motif family, member 2 |
| chr17_-_80797886 | 0.55 |
ENST00000572562.1
|
ZNF750
|
zinc finger protein 750 |
| chr17_+_75401152 | 0.54 |
ENST00000585930.1
|
SEPT9
|
septin 9 |
| chr12_+_41136144 | 0.54 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr5_-_142814241 | 0.53 |
ENST00000504572.1
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr7_+_56032652 | 0.52 |
ENST00000437587.1
|
GBAS
|
glioblastoma amplified sequence |
| chr8_-_49834299 | 0.52 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr17_-_8059638 | 0.51 |
ENST00000584202.1
ENST00000354903.5 ENST00000577253.1 |
PER1
|
period circadian clock 1 |
| chrX_+_153029633 | 0.51 |
ENST00000538966.1
ENST00000361971.5 ENST00000538776.1 ENST00000538543.1 |
PLXNB3
|
plexin B3 |
| chr11_+_57435441 | 0.50 |
ENST00000528177.1
|
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr6_-_32634425 | 0.50 |
ENST00000399082.3
ENST00000399079.3 ENST00000374943.4 ENST00000434651.2 |
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1 |
| chr10_+_116697946 | 0.50 |
ENST00000298746.3
|
TRUB1
|
TruB pseudouridine (psi) synthase family member 1 |
| chr7_-_38948774 | 0.50 |
ENST00000395969.2
ENST00000414632.1 ENST00000310301.4 |
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr7_-_130353553 | 0.50 |
ENST00000330992.7
ENST00000445977.2 |
COPG2
|
coatomer protein complex, subunit gamma 2 |
| chr9_+_71820057 | 0.50 |
ENST00000539225.1
|
TJP2
|
tight junction protein 2 |
| chr1_-_46642154 | 0.49 |
ENST00000540385.1
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr11_+_64950801 | 0.48 |
ENST00000526468.1
|
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr1_-_31538517 | 0.48 |
ENST00000440538.2
ENST00000423018.2 ENST00000424085.2 ENST00000426105.2 ENST00000257075.5 ENST00000373747.3 ENST00000525843.1 ENST00000373742.2 |
PUM1
|
pumilio RNA-binding family member 1 |
| chr2_-_37501692 | 0.47 |
ENST00000443977.1
|
PRKD3
|
protein kinase D3 |
| chr5_+_138210919 | 0.47 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr8_-_49833978 | 0.47 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr22_-_30814469 | 0.47 |
ENST00000598426.1
|
KIAA1658
|
KIAA1658 |
| chr12_+_14572070 | 0.47 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr11_+_33563618 | 0.46 |
ENST00000526400.1
|
KIAA1549L
|
KIAA1549-like |
| chr6_-_111927062 | 0.46 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr18_-_43684230 | 0.46 |
ENST00000592989.1
ENST00000589869.1 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr9_+_71819927 | 0.46 |
ENST00000535702.1
|
TJP2
|
tight junction protein 2 |
| chr13_-_30881134 | 0.45 |
ENST00000380617.3
ENST00000441394.1 |
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr3_+_186743261 | 0.44 |
ENST00000423451.1
ENST00000446170.1 |
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr1_-_35325318 | 0.44 |
ENST00000423898.1
ENST00000456842.1 |
SMIM12
|
small integral membrane protein 12 |
| chr7_+_134430212 | 0.43 |
ENST00000436461.2
|
CALD1
|
caldesmon 1 |
| chr5_-_110848253 | 0.43 |
ENST00000505803.1
ENST00000502322.1 |
STARD4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr2_-_31440377 | 0.43 |
ENST00000444918.2
ENST00000403897.3 |
CAPN14
|
calpain 14 |
| chr4_-_90757364 | 0.43 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr3_-_69062742 | 0.42 |
ENST00000424374.1
|
EOGT
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chrX_-_108976410 | 0.42 |
ENST00000504980.1
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4 |
| chr2_+_201170596 | 0.42 |
ENST00000439084.1
ENST00000409718.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr4_+_146403912 | 0.41 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr4_-_90758118 | 0.41 |
ENST00000420646.2
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr14_+_64565442 | 0.41 |
ENST00000553308.1
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr14_-_91720224 | 0.40 |
ENST00000238699.3
ENST00000531499.2 |
GPR68
|
G protein-coupled receptor 68 |
| chr2_-_172291273 | 0.39 |
ENST00000442778.1
ENST00000453846.1 |
METTL8
|
methyltransferase like 8 |
| chr15_+_40674920 | 0.38 |
ENST00000416151.2
ENST00000249776.8 |
KNSTRN
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr12_-_123565834 | 0.38 |
ENST00000546049.1
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chrX_+_69509870 | 0.38 |
ENST00000374388.3
|
KIF4A
|
kinesin family member 4A |
| chr16_-_18911366 | 0.38 |
ENST00000565224.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr3_-_122233723 | 0.38 |
ENST00000493510.1
ENST00000344337.6 ENST00000476916.1 ENST00000465882.1 |
KPNA1
|
karyopherin alpha 1 (importin alpha 5) |
| chr5_+_108083517 | 0.37 |
ENST00000281092.4
ENST00000536402.1 |
FER
|
fer (fps/fes related) tyrosine kinase |
| chr2_-_38303218 | 0.37 |
ENST00000407341.1
ENST00000260630.3 |
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1 |
| chr3_-_123512688 | 0.36 |
ENST00000475616.1
|
MYLK
|
myosin light chain kinase |
| chr2_+_172309634 | 0.36 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr10_+_1120312 | 0.36 |
ENST00000436154.1
|
WDR37
|
WD repeat domain 37 |
| chr2_+_64069240 | 0.35 |
ENST00000497883.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr3_+_182971018 | 0.35 |
ENST00000326505.3
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr3_+_30647994 | 0.35 |
ENST00000295754.5
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chrX_-_134049233 | 0.35 |
ENST00000370779.4
|
MOSPD1
|
motile sperm domain containing 1 |
| chrX_+_9502971 | 0.35 |
ENST00000452824.1
|
TBL1X
|
transducin (beta)-like 1X-linked |
| chr10_+_99332198 | 0.35 |
ENST00000307518.5
ENST00000298808.5 ENST00000370655.1 |
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr14_+_36295638 | 0.34 |
ENST00000543183.1
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr17_+_26662597 | 0.34 |
ENST00000544907.2
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr14_+_100531738 | 0.33 |
ENST00000555706.1
|
EVL
|
Enah/Vasp-like |
| chr4_-_36246060 | 0.33 |
ENST00000303965.4
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr2_+_71357744 | 0.33 |
ENST00000498451.2
|
MPHOSPH10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr3_-_69062764 | 0.32 |
ENST00000295571.5
|
EOGT
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr6_-_32160622 | 0.32 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chr8_-_99954788 | 0.32 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr5_+_49961727 | 0.32 |
ENST00000505697.2
ENST00000503750.2 ENST00000514342.2 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr2_+_64069016 | 0.31 |
ENST00000488245.2
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr15_-_78369994 | 0.31 |
ENST00000300584.3
ENST00000409931.3 |
TBC1D2B
|
TBC1 domain family, member 2B |
| chr22_+_21213771 | 0.31 |
ENST00000439214.1
|
SNAP29
|
synaptosomal-associated protein, 29kDa |
| chr10_-_104262460 | 0.31 |
ENST00000446605.2
ENST00000369905.4 ENST00000545684.1 |
ACTR1A
|
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
| chr7_+_7196565 | 0.31 |
ENST00000429911.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr1_-_89488510 | 0.31 |
ENST00000564665.1
ENST00000370481.4 |
GBP3
|
guanylate binding protein 3 |
| chr10_+_13203543 | 0.30 |
ENST00000378714.3
ENST00000479669.1 ENST00000484800.2 |
MCM10
|
minichromosome maintenance complex component 10 |
| chr1_+_174670143 | 0.30 |
ENST00000367687.1
ENST00000347255.2 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr1_-_68962805 | 0.30 |
ENST00000370966.5
|
DEPDC1
|
DEP domain containing 1 |
| chr9_+_131644398 | 0.30 |
ENST00000372599.3
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr2_+_219110149 | 0.30 |
ENST00000456575.1
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr19_+_45409011 | 0.30 |
ENST00000252486.4
ENST00000446996.1 ENST00000434152.1 |
APOE
|
apolipoprotein E |
| chr1_-_232598163 | 0.29 |
ENST00000308942.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr1_-_200379104 | 0.29 |
ENST00000367352.3
|
ZNF281
|
zinc finger protein 281 |
| chr1_+_65613340 | 0.29 |
ENST00000546702.1
|
AK4
|
adenylate kinase 4 |
| chr19_+_53761545 | 0.28 |
ENST00000341702.3
|
VN1R2
|
vomeronasal 1 receptor 2 |
| chr10_-_36813162 | 0.28 |
ENST00000440465.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr3_+_101818088 | 0.28 |
ENST00000491959.1
|
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr9_+_131644388 | 0.28 |
ENST00000372600.4
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr15_+_40674963 | 0.27 |
ENST00000448395.2
|
KNSTRN
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr17_-_8055747 | 0.27 |
ENST00000317276.4
ENST00000581703.1 |
PER1
|
period circadian clock 1 |
| chr13_-_30880979 | 0.27 |
ENST00000414289.1
|
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr10_-_73976884 | 0.27 |
ENST00000317126.4
ENST00000545550.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr16_-_58663720 | 0.27 |
ENST00000564557.1
ENST00000569240.1 ENST00000441024.2 ENST00000569020.1 ENST00000317147.5 |
CNOT1
|
CCR4-NOT transcription complex, subunit 1 |
| chr3_+_39093481 | 0.26 |
ENST00000302313.5
ENST00000544962.1 ENST00000396258.3 ENST00000418020.1 |
WDR48
|
WD repeat domain 48 |
| chr2_+_232135245 | 0.26 |
ENST00000446447.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr18_-_43684186 | 0.26 |
ENST00000590406.1
ENST00000282050.2 ENST00000590324.1 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr4_-_39034542 | 0.26 |
ENST00000344606.6
|
TMEM156
|
transmembrane protein 156 |
| chr19_+_14492217 | 0.26 |
ENST00000587606.1
ENST00000586517.1 ENST00000591080.1 |
CD97
|
CD97 molecule |
| chr2_+_64068844 | 0.25 |
ENST00000337130.5
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr4_-_141677267 | 0.25 |
ENST00000442267.2
|
TBC1D9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr1_-_35325400 | 0.25 |
ENST00000521580.2
|
SMIM12
|
small integral membrane protein 12 |
| chrX_-_40036520 | 0.25 |
ENST00000406200.2
ENST00000378455.4 ENST00000342274.4 |
BCOR
|
BCL6 corepressor |
| chr11_-_71752838 | 0.25 |
ENST00000537930.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr11_+_7595136 | 0.24 |
ENST00000529575.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr12_+_6309963 | 0.24 |
ENST00000382515.2
|
CD9
|
CD9 molecule |
| chr8_+_75262629 | 0.24 |
ENST00000434412.2
|
GDAP1
|
ganglioside induced differentiation associated protein 1 |
| chr10_+_129785536 | 0.24 |
ENST00000419012.2
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chr14_-_94547541 | 0.24 |
ENST00000544005.1
ENST00000555054.1 ENST00000330836.5 |
DDX24
|
DEAD (Asp-Glu-Ala-Asp) box helicase 24 |
| chr2_+_64069459 | 0.24 |
ENST00000445915.2
ENST00000475462.1 |
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr5_-_39270725 | 0.24 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr10_-_104913367 | 0.24 |
ENST00000423468.2
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr8_-_135652051 | 0.24 |
ENST00000522257.1
|
ZFAT
|
zinc finger and AT hook domain containing |
| chr19_-_10341948 | 0.24 |
ENST00000590320.1
ENST00000592342.1 ENST00000588952.1 |
S1PR2
DNMT1
|
sphingosine-1-phosphate receptor 2 DNA (cytosine-5-)-methyltransferase 1 |
| chr10_-_27529486 | 0.23 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr1_+_46806452 | 0.23 |
ENST00000536062.1
|
NSUN4
|
NOP2/Sun domain family, member 4 |
| chr13_-_50018241 | 0.23 |
ENST00000409308.1
|
CAB39L
|
calcium binding protein 39-like |
| chr12_+_12966250 | 0.23 |
ENST00000352940.4
ENST00000358007.3 ENST00000544400.1 |
DDX47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr11_+_12399071 | 0.23 |
ENST00000539723.1
ENST00000550549.1 |
PARVA
|
parvin, alpha |
| chr10_+_27793197 | 0.23 |
ENST00000356940.6
ENST00000535776.1 |
RAB18
|
RAB18, member RAS oncogene family |
| chr3_+_30648066 | 0.22 |
ENST00000359013.4
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr1_+_110091189 | 0.22 |
ENST00000369851.4
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr2_-_36825281 | 0.22 |
ENST00000405912.3
ENST00000379245.4 |
FEZ2
|
fasciculation and elongation protein zeta 2 (zygin II) |
| chr13_-_50018140 | 0.22 |
ENST00000410043.1
ENST00000347776.5 |
CAB39L
|
calcium binding protein 39-like |
| chr1_+_46806837 | 0.22 |
ENST00000537428.1
|
NSUN4
|
NOP2/Sun domain family, member 4 |
| chrX_+_109602039 | 0.22 |
ENST00000520821.1
|
RGAG1
|
retrotransposon gag domain containing 1 |
| chrX_+_37865804 | 0.22 |
ENST00000297875.2
ENST00000357972.5 |
SYTL5
|
synaptotagmin-like 5 |
| chr2_-_54197915 | 0.21 |
ENST00000404125.1
|
PSME4
|
proteasome (prosome, macropain) activator subunit 4 |
| chr10_+_99332529 | 0.21 |
ENST00000455090.1
|
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr4_+_144303093 | 0.21 |
ENST00000505913.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr15_+_43477455 | 0.21 |
ENST00000300213.4
|
CCNDBP1
|
cyclin D-type binding-protein 1 |
| chr14_-_23762777 | 0.20 |
ENST00000431326.2
|
HOMEZ
|
homeobox and leucine zipper encoding |
| chr12_+_19389814 | 0.20 |
ENST00000536974.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr10_+_129785574 | 0.20 |
ENST00000430713.2
ENST00000471218.1 |
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chr19_+_14491948 | 0.20 |
ENST00000358600.3
|
CD97
|
CD97 molecule |
| chr2_+_102803433 | 0.19 |
ENST00000264257.2
ENST00000441515.2 |
IL1RL2
|
interleukin 1 receptor-like 2 |
| chr15_+_43477580 | 0.19 |
ENST00000356633.5
|
CCNDBP1
|
cyclin D-type binding-protein 1 |
| chr3_-_42623805 | 0.19 |
ENST00000456515.1
|
SEC22C
|
SEC22 vesicle trafficking protein homolog C (S. cerevisiae) |
| chr1_+_174669653 | 0.19 |
ENST00000325589.5
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chrX_-_16730984 | 0.19 |
ENST00000380241.3
|
CTPS2
|
CTP synthase 2 |
| chr1_+_109632425 | 0.19 |
ENST00000338272.8
|
TMEM167B
|
transmembrane protein 167B |
| chr11_+_11863500 | 0.19 |
ENST00000527733.1
ENST00000539466.1 |
USP47
|
ubiquitin specific peptidase 47 |
| chr14_+_36295504 | 0.18 |
ENST00000216807.7
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr1_+_40839369 | 0.18 |
ENST00000372718.3
|
SMAP2
|
small ArfGAP2 |
| chr11_-_71753188 | 0.18 |
ENST00000543009.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr19_-_19051993 | 0.18 |
ENST00000594794.1
ENST00000355887.6 ENST00000392351.3 ENST00000596482.1 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr2_+_161993465 | 0.18 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chrX_-_32173579 | 0.18 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr3_+_183892635 | 0.17 |
ENST00000427072.1
ENST00000411763.2 ENST00000292807.5 ENST00000448139.1 ENST00000455925.1 |
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr16_+_68877496 | 0.17 |
ENST00000261778.1
|
TANGO6
|
transport and golgi organization 6 homolog (Drosophila) |
| chr5_-_42887494 | 0.17 |
ENST00000514218.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr2_+_114163945 | 0.17 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr15_+_33022885 | 0.17 |
ENST00000322805.4
|
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr10_+_3146868 | 0.17 |
ENST00000415005.1
ENST00000468050.1 |
PFKP
|
phosphofructokinase, platelet |
| chr1_-_182573514 | 0.17 |
ENST00000367558.5
|
RGS16
|
regulator of G-protein signaling 16 |
| chr6_+_46761118 | 0.17 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr6_-_150212029 | 0.16 |
ENST00000529948.1
ENST00000357183.4 ENST00000367363.3 |
RAET1E
|
retinoic acid early transcript 1E |
| chr3_+_38179969 | 0.16 |
ENST00000396334.3
ENST00000417037.2 ENST00000424893.1 ENST00000495303.1 ENST00000443433.2 ENST00000421516.1 |
MYD88
|
myeloid differentiation primary response 88 |
| chr6_-_33239712 | 0.16 |
ENST00000436044.2
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr19_+_50084561 | 0.16 |
ENST00000246794.5
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr14_+_55494323 | 0.16 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr14_+_67831576 | 0.16 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr20_+_56964169 | 0.16 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr16_+_56969284 | 0.16 |
ENST00000568358.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr22_+_45714301 | 0.16 |
ENST00000427777.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr10_+_16478942 | 0.16 |
ENST00000535784.2
ENST00000423462.2 ENST00000378000.1 |
PTER
|
phosphotriesterase related |
| chr12_-_122018859 | 0.16 |
ENST00000536437.1
ENST00000377071.4 ENST00000538046.2 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr3_+_190105909 | 0.16 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr15_-_99791413 | 0.15 |
ENST00000394129.2
ENST00000558663.1 ENST00000394135.3 ENST00000561365.1 ENST00000560279.1 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr1_+_152730499 | 0.15 |
ENST00000368773.1
|
KPRP
|
keratinocyte proline-rich protein |
| chr9_+_135906076 | 0.15 |
ENST00000372097.5
ENST00000440319.1 |
GTF3C5
|
general transcription factor IIIC, polypeptide 5, 63kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0051620 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.3 | 1.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.3 | 2.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.3 | 0.9 | GO:0010877 | lipid transport involved in lipid storage(GO:0010877) |
| 0.2 | 1.8 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.2 | 0.6 | GO:0002661 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.2 | 1.5 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.2 | 0.8 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.6 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 0.5 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.1 | 0.4 | GO:0050904 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) diapedesis(GO:0050904) |
| 0.1 | 0.7 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.7 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 1.3 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.1 | 0.6 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 | 0.8 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 0.7 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 1.0 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 0.5 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.4 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 | 0.2 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 | 0.6 | GO:1903788 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.1 | 0.3 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 0.7 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.5 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.8 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.3 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 0.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.2 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.1 | 0.5 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.6 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 0.2 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.1 | 0.4 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.5 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.5 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.6 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.1 | GO:1903598 | positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.5 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.1 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.0 | 0.4 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.4 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.4 | GO:1902365 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.2 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 | 0.4 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.5 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.8 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.5 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.8 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.2 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.3 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.0 | 0.7 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.5 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 1.0 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.2 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.6 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.4 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.7 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.2 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.3 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.2 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.0 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.1 | GO:2000691 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.5 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.5 | GO:0001783 | B cell apoptotic process(GO:0001783) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.6 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.3 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.5 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.4 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.9 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.0 | 0.1 | GO:0019249 | regulation of amino acid import(GO:0010958) lactate biosynthetic process(GO:0019249) regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.3 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.0 | 0.1 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.0 | GO:0090472 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 1.7 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.6 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.1 | 0.3 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.3 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.7 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 1.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.3 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.7 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) glial limiting end-foot(GO:0097451) |
| 0.1 | 0.6 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.3 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.1 | 0.5 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 2.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.4 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.3 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.4 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.5 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 1.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.5 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.3 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.3 | 1.8 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.2 | 1.4 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.2 | 0.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 0.6 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 0.6 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.7 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.1 | 0.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.6 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.5 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.1 | 0.7 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 2.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.8 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.3 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.2 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.1 | 0.3 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.1 | 0.3 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 0.5 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 0.4 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.4 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.5 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.2 | GO:0001003 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.6 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 1.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.1 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.6 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 1.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.0 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.7 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 2.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.8 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.5 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |