Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
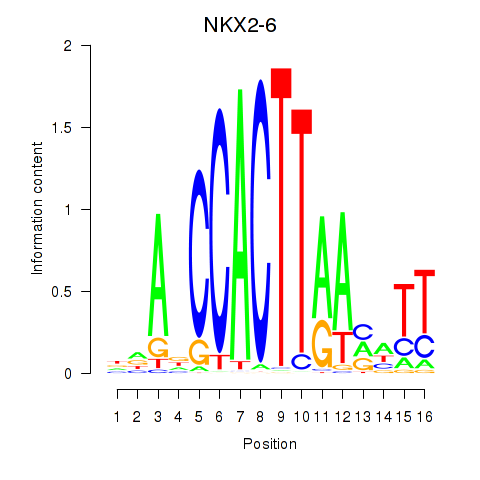
Results for NKX2-6
Z-value: 0.68
Transcription factors associated with NKX2-6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-6
|
ENSG00000180053.6 | NK2 homeobox 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-6 | hg19_v2_chr8_-_23563922_23564111 | 0.04 | 8.6e-01 | Click! |
Activity profile of NKX2-6 motif
Sorted Z-values of NKX2-6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_120365866 | 2.21 |
ENST00000475447.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr12_+_56075330 | 1.94 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr5_-_59481406 | 1.58 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr8_+_27629459 | 1.56 |
ENST00000523566.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr15_-_56757329 | 1.31 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr9_-_19065082 | 1.27 |
ENST00000415524.1
|
HAUS6
|
HAUS augmin-like complex, subunit 6 |
| chr11_-_61687739 | 1.27 |
ENST00000531922.1
ENST00000301773.5 |
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr7_-_107880508 | 1.16 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
| chr9_+_97766409 | 1.11 |
ENST00000425634.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr8_+_99076509 | 1.09 |
ENST00000318528.3
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr1_-_36863481 | 1.05 |
ENST00000315732.2
|
LSM10
|
LSM10, U7 small nuclear RNA associated |
| chr17_-_57229155 | 1.04 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr8_+_41386761 | 1.04 |
ENST00000523277.2
|
GINS4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr12_+_113354341 | 1.01 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr22_+_44427230 | 0.97 |
ENST00000444029.1
|
PARVB
|
parvin, beta |
| chr8_-_95220775 | 0.97 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr20_-_32262165 | 0.96 |
ENST00000606690.1
ENST00000246190.6 ENST00000439478.1 ENST00000375238.4 |
NECAB3
|
N-terminal EF-hand calcium binding protein 3 |
| chr12_-_8088773 | 0.94 |
ENST00000544291.1
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr3_-_123339343 | 0.94 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr15_+_65843130 | 0.88 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr20_-_5100591 | 0.87 |
ENST00000379143.5
|
PCNA
|
proliferating cell nuclear antigen |
| chr12_-_8088871 | 0.87 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr8_+_42396712 | 0.81 |
ENST00000518574.1
ENST00000417410.2 ENST00000414154.2 |
SMIM19
|
small integral membrane protein 19 |
| chr10_+_135204338 | 0.80 |
ENST00000468317.2
|
RP11-108K14.8
|
Mitochondrial GTPase 1 |
| chr17_-_34890037 | 0.80 |
ENST00000589404.1
|
MYO19
|
myosin XIX |
| chr17_-_39341594 | 0.78 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr17_+_49337881 | 0.76 |
ENST00000225298.7
|
UTP18
|
UTP18 small subunit (SSU) processome component homolog (yeast) |
| chr2_-_121624973 | 0.75 |
ENST00000603720.1
|
RP11-297J22.1
|
RP11-297J22.1 |
| chr20_-_271009 | 0.70 |
ENST00000382369.5
|
C20orf96
|
chromosome 20 open reading frame 96 |
| chr3_-_123339418 | 0.69 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr3_+_37035289 | 0.68 |
ENST00000455445.2
ENST00000441265.1 ENST00000435176.1 ENST00000429117.1 ENST00000536378.1 |
MLH1
|
mutL homolog 1 |
| chr17_-_34890665 | 0.68 |
ENST00000586007.1
|
MYO19
|
myosin XIX |
| chr11_+_65627974 | 0.67 |
ENST00000525768.1
|
MUS81
|
MUS81 structure-specific endonuclease subunit |
| chr7_-_56101826 | 0.66 |
ENST00000421626.1
|
PSPH
|
phosphoserine phosphatase |
| chr11_+_118958689 | 0.66 |
ENST00000535253.1
ENST00000392841.1 |
HMBS
|
hydroxymethylbilane synthase |
| chr3_-_127541194 | 0.64 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chr19_-_51611623 | 0.63 |
ENST00000421832.2
|
CTU1
|
cytosolic thiouridylase subunit 1 |
| chr17_-_44896047 | 0.61 |
ENST00000225512.5
|
WNT3
|
wingless-type MMTV integration site family, member 3 |
| chr2_+_87769459 | 0.60 |
ENST00000414030.1
ENST00000437561.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr15_+_84841242 | 0.60 |
ENST00000558195.1
|
RP11-182J1.16
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr17_-_34890617 | 0.59 |
ENST00000586886.1
ENST00000585719.1 ENST00000585818.1 |
MYO19
|
myosin XIX |
| chr6_+_126102292 | 0.57 |
ENST00000368357.3
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr17_+_33914276 | 0.57 |
ENST00000592545.1
ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr17_-_34890709 | 0.56 |
ENST00000544606.1
|
MYO19
|
myosin XIX |
| chr12_-_89919965 | 0.56 |
ENST00000548729.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr19_-_49552363 | 0.56 |
ENST00000448456.3
ENST00000355414.2 |
CGB8
|
chorionic gonadotropin, beta polypeptide 8 |
| chr7_-_37382360 | 0.55 |
ENST00000455119.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr8_+_42396936 | 0.55 |
ENST00000416469.2
|
SMIM19
|
small integral membrane protein 19 |
| chr18_+_68002675 | 0.55 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr19_-_49843539 | 0.55 |
ENST00000602554.1
ENST00000358234.4 |
CTC-301O7.4
|
CTC-301O7.4 |
| chr15_-_72668805 | 0.54 |
ENST00000268097.5
|
HEXA
|
hexosaminidase A (alpha polypeptide) |
| chr11_-_118901559 | 0.54 |
ENST00000330775.7
ENST00000545985.1 ENST00000357590.5 ENST00000538950.1 |
SLC37A4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chrX_-_53310791 | 0.54 |
ENST00000375365.2
|
IQSEC2
|
IQ motif and Sec7 domain 2 |
| chr8_-_62602327 | 0.53 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr3_-_157824292 | 0.52 |
ENST00000483851.2
|
SHOX2
|
short stature homeobox 2 |
| chr17_+_33914460 | 0.51 |
ENST00000537622.2
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr10_-_27529486 | 0.50 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr16_-_69373396 | 0.50 |
ENST00000562595.1
ENST00000562081.1 ENST00000306875.4 |
COG8
|
component of oligomeric golgi complex 8 |
| chr17_+_41150290 | 0.50 |
ENST00000589037.1
ENST00000253788.5 |
RPL27
|
ribosomal protein L27 |
| chr12_-_89920030 | 0.50 |
ENST00000413530.1
ENST00000547474.1 |
GALNT4
POC1B-GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr3_-_114790179 | 0.50 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr21_-_46964306 | 0.50 |
ENST00000443742.1
ENST00000528477.1 ENST00000567670.1 |
SLC19A1
|
solute carrier family 19 (folate transporter), member 1 |
| chr17_-_62493131 | 0.49 |
ENST00000539111.2
|
POLG2
|
polymerase (DNA directed), gamma 2, accessory subunit |
| chr14_-_30766223 | 0.48 |
ENST00000549360.1
ENST00000508469.2 |
CTD-2251F13.1
|
CTD-2251F13.1 |
| chr7_-_150777874 | 0.47 |
ENST00000540185.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr7_-_150777920 | 0.47 |
ENST00000353841.2
ENST00000297532.6 |
FASTK
|
Fas-activated serine/threonine kinase |
| chr8_+_22414182 | 0.47 |
ENST00000524057.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr10_-_13342097 | 0.46 |
ENST00000263038.4
|
PHYH
|
phytanoyl-CoA 2-hydroxylase |
| chr14_+_50779029 | 0.46 |
ENST00000245448.6
|
ATP5S
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr2_-_175462934 | 0.46 |
ENST00000392546.2
ENST00000436221.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr15_-_55563072 | 0.46 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr12_-_89919765 | 0.45 |
ENST00000541909.1
ENST00000313546.3 |
POC1B
|
POC1 centriolar protein B |
| chr5_+_52083730 | 0.45 |
ENST00000282588.6
ENST00000274311.2 |
ITGA1
PELO
|
integrin, alpha 1 pelota homolog (Drosophila) |
| chr4_-_129491686 | 0.45 |
ENST00000514265.1
|
RP11-184M15.1
|
RP11-184M15.1 |
| chr6_+_24775641 | 0.44 |
ENST00000378054.2
ENST00000476555.1 |
GMNN
|
geminin, DNA replication inhibitor |
| chr17_+_60501228 | 0.43 |
ENST00000311506.5
|
METTL2A
|
methyltransferase like 2A |
| chr17_+_33914424 | 0.43 |
ENST00000590432.1
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr17_+_34890807 | 0.43 |
ENST00000429467.2
ENST00000592983.1 |
PIGW
|
phosphatidylinositol glycan anchor biosynthesis, class W |
| chr15_-_65579177 | 0.42 |
ENST00000444347.2
ENST00000261888.6 |
PARP16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr11_+_65627865 | 0.42 |
ENST00000308110.4
|
MUS81
|
MUS81 structure-specific endonuclease subunit |
| chr17_-_6524159 | 0.42 |
ENST00000589033.1
|
KIAA0753
|
KIAA0753 |
| chr9_-_111882195 | 0.41 |
ENST00000374586.3
|
TMEM245
|
transmembrane protein 245 |
| chr1_+_173793641 | 0.40 |
ENST00000361951.4
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr2_-_175462456 | 0.40 |
ENST00000409891.1
ENST00000410117.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr14_+_31494841 | 0.40 |
ENST00000556232.1
ENST00000216366.4 ENST00000334725.4 ENST00000554609.1 ENST00000554345.1 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr7_-_150777949 | 0.40 |
ENST00000482571.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr10_+_121652204 | 0.40 |
ENST00000369075.3
ENST00000543134.1 |
SEC23IP
|
SEC23 interacting protein |
| chr8_-_121457608 | 0.40 |
ENST00000306185.3
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr12_-_57039739 | 0.39 |
ENST00000552959.1
ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr12_+_118454500 | 0.39 |
ENST00000537315.1
ENST00000229043.3 ENST00000484086.2 ENST00000420967.1 ENST00000454402.2 ENST00000392542.2 ENST00000535092.1 |
RFC5
|
replication factor C (activator 1) 5, 36.5kDa |
| chr14_+_31494672 | 0.39 |
ENST00000542754.2
ENST00000313566.6 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr1_-_173793246 | 0.39 |
ENST00000345664.6
ENST00000367710.3 |
CENPL
|
centromere protein L |
| chr6_-_131299929 | 0.39 |
ENST00000531356.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr17_+_41150793 | 0.38 |
ENST00000586277.1
|
RPL27
|
ribosomal protein L27 |
| chr17_+_58755184 | 0.38 |
ENST00000589222.1
ENST00000407086.3 ENST00000390652.5 |
BCAS3
|
breast carcinoma amplified sequence 3 |
| chr9_-_4859260 | 0.38 |
ENST00000599351.1
|
AL158147.2
|
HCG2011465; Uncharacterized protein |
| chr4_+_113739244 | 0.38 |
ENST00000503271.1
ENST00000503423.1 ENST00000506722.1 |
ANK2
|
ankyrin 2, neuronal |
| chr1_+_110162448 | 0.37 |
ENST00000342115.4
ENST00000469039.2 ENST00000474459.1 ENST00000528667.1 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr14_-_31856397 | 0.37 |
ENST00000538864.2
ENST00000550366.1 |
HEATR5A
|
HEAT repeat containing 5A |
| chr3_-_39196049 | 0.36 |
ENST00000514182.1
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1 |
| chrX_-_47518527 | 0.35 |
ENST00000333119.3
|
UXT
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr20_-_36793663 | 0.34 |
ENST00000536701.1
ENST00000536724.1 |
TGM2
|
transglutaminase 2 |
| chr2_-_69180083 | 0.34 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chr4_-_170924888 | 0.34 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr1_-_149785236 | 0.34 |
ENST00000331491.1
|
HIST2H3D
|
histone cluster 2, H3d |
| chr12_-_109531264 | 0.34 |
ENST00000429722.2
ENST00000536242.1 ENST00000343075.3 ENST00000536358.1 |
ALKBH2
|
alkB, alkylation repair homolog 2 (E. coli) |
| chr17_-_34890732 | 0.33 |
ENST00000268852.9
|
MYO19
|
myosin XIX |
| chr8_+_99076750 | 0.33 |
ENST00000545282.1
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr19_+_42746927 | 0.33 |
ENST00000378108.1
|
AC006486.1
|
AC006486.1 |
| chr9_+_116263639 | 0.33 |
ENST00000343817.5
|
RGS3
|
regulator of G-protein signaling 3 |
| chr1_-_160001737 | 0.33 |
ENST00000368090.2
|
PIGM
|
phosphatidylinositol glycan anchor biosynthesis, class M |
| chr1_+_173793777 | 0.33 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr17_-_41116454 | 0.32 |
ENST00000427569.2
ENST00000430739.1 |
AARSD1
|
alanyl-tRNA synthetase domain containing 1 |
| chr13_-_26452679 | 0.31 |
ENST00000596729.1
|
AL138815.2
|
Uncharacterized protein |
| chrX_-_47518498 | 0.31 |
ENST00000335890.2
|
UXT
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr2_-_202298268 | 0.31 |
ENST00000440597.1
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr9_+_134065506 | 0.31 |
ENST00000483497.2
|
NUP214
|
nucleoporin 214kDa |
| chr14_-_60337684 | 0.30 |
ENST00000267484.5
|
RTN1
|
reticulon 1 |
| chr17_-_47723943 | 0.30 |
ENST00000510476.1
ENST00000503676.1 |
SPOP
|
speckle-type POZ protein |
| chr6_+_10528560 | 0.30 |
ENST00000379597.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr17_-_34890759 | 0.30 |
ENST00000431794.3
|
MYO19
|
myosin XIX |
| chr1_-_115259337 | 0.30 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr1_-_173793458 | 0.30 |
ENST00000356198.2
|
CENPL
|
centromere protein L |
| chr2_+_219246746 | 0.29 |
ENST00000233202.6
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr14_+_50779071 | 0.29 |
ENST00000426751.2
|
ATP5S
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr16_+_69373323 | 0.28 |
ENST00000254940.5
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr3_-_160167301 | 0.28 |
ENST00000494486.1
|
TRIM59
|
tripartite motif containing 59 |
| chr12_+_120031264 | 0.28 |
ENST00000426426.1
|
TMEM233
|
transmembrane protein 233 |
| chr19_-_44174330 | 0.27 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr3_+_37034823 | 0.27 |
ENST00000231790.2
ENST00000456676.2 |
MLH1
|
mutL homolog 1 |
| chr2_-_36779411 | 0.26 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr1_-_200638964 | 0.26 |
ENST00000367348.3
ENST00000447706.2 ENST00000331314.6 |
DDX59
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 59 |
| chr11_+_94277017 | 0.26 |
ENST00000358752.2
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr11_-_64527425 | 0.25 |
ENST00000377432.3
|
PYGM
|
phosphorylase, glycogen, muscle |
| chr12_-_39300071 | 0.25 |
ENST00000550863.1
|
CPNE8
|
copine VIII |
| chr11_-_76381029 | 0.25 |
ENST00000407242.2
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr8_-_82644562 | 0.24 |
ENST00000520604.1
ENST00000521742.1 ENST00000520635.1 |
ZFAND1
|
zinc finger, AN1-type domain 1 |
| chr9_-_134406611 | 0.24 |
ENST00000372208.3
ENST00000372215.4 |
UCK1
|
uridine-cytidine kinase 1 |
| chr9_+_134065519 | 0.24 |
ENST00000531600.1
|
NUP214
|
nucleoporin 214kDa |
| chr13_-_88463487 | 0.24 |
ENST00000606221.1
|
RP11-471M2.3
|
RP11-471M2.3 |
| chr6_-_144416737 | 0.24 |
ENST00000367569.2
|
SF3B5
|
splicing factor 3b, subunit 5, 10kDa |
| chr9_-_70465758 | 0.24 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr17_-_74049720 | 0.24 |
ENST00000602720.1
|
SRP68
|
signal recognition particle 68kDa |
| chr10_-_91403625 | 0.23 |
ENST00000322191.6
ENST00000342512.3 ENST00000371774.2 |
PANK1
|
pantothenate kinase 1 |
| chr3_+_32726620 | 0.23 |
ENST00000331889.6
ENST00000328834.5 |
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr19_+_56111680 | 0.23 |
ENST00000301073.3
|
ZNF524
|
zinc finger protein 524 |
| chr19_+_52076425 | 0.22 |
ENST00000436511.2
|
ZNF175
|
zinc finger protein 175 |
| chr5_-_139937895 | 0.22 |
ENST00000336283.6
|
SRA1
|
steroid receptor RNA activator 1 |
| chr3_+_140981456 | 0.22 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr3_-_81811312 | 0.22 |
ENST00000429644.2
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr22_+_42017987 | 0.22 |
ENST00000405506.1
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr16_+_69796209 | 0.22 |
ENST00000359154.2
ENST00000561780.1 ENST00000563659.1 ENST00000448661.1 |
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr7_+_16700806 | 0.21 |
ENST00000446596.1
ENST00000438834.1 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr13_+_27998681 | 0.21 |
ENST00000381140.4
|
GTF3A
|
general transcription factor IIIA |
| chr17_-_7216939 | 0.21 |
ENST00000573684.1
|
GPS2
|
G protein pathway suppressor 2 |
| chr4_+_169418195 | 0.21 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr1_+_110163202 | 0.20 |
ENST00000531203.1
ENST00000256578.3 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr12_+_44152740 | 0.20 |
ENST00000440781.2
ENST00000431837.1 ENST00000550616.1 ENST00000448290.2 ENST00000551736.1 |
IRAK4
|
interleukin-1 receptor-associated kinase 4 |
| chr15_+_43425672 | 0.20 |
ENST00000260403.2
|
TMEM62
|
transmembrane protein 62 |
| chr1_+_171217677 | 0.20 |
ENST00000402921.2
|
FMO1
|
flavin containing monooxygenase 1 |
| chr1_+_159796534 | 0.20 |
ENST00000289707.5
|
SLAMF8
|
SLAM family member 8 |
| chr9_-_33402506 | 0.20 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr19_+_3178736 | 0.19 |
ENST00000246115.3
|
S1PR4
|
sphingosine-1-phosphate receptor 4 |
| chr2_+_86426478 | 0.18 |
ENST00000254644.8
ENST00000605125.1 ENST00000337109.4 ENST00000409180.1 |
MRPL35
|
mitochondrial ribosomal protein L35 |
| chr19_-_44174305 | 0.18 |
ENST00000601723.1
ENST00000339082.3 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr5_+_140071011 | 0.18 |
ENST00000230771.3
ENST00000509299.1 ENST00000503873.1 ENST00000435019.2 ENST00000437649.2 ENST00000432671.2 |
HARS2
|
histidyl-tRNA synthetase 2, mitochondrial |
| chr10_+_32735030 | 0.18 |
ENST00000277657.6
ENST00000362006.5 |
CCDC7
|
coiled-coil domain containing 7 |
| chr6_-_15586238 | 0.18 |
ENST00000462989.2
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr2_-_220252530 | 0.18 |
ENST00000521459.1
|
DNPEP
|
aspartyl aminopeptidase |
| chr12_-_68845165 | 0.18 |
ENST00000360485.3
ENST00000441255.2 |
RP11-81H14.2
|
RP11-81H14.2 |
| chr3_+_156807663 | 0.17 |
ENST00000467995.1
ENST00000474477.1 ENST00000471719.1 |
LINC00881
|
long intergenic non-protein coding RNA 881 |
| chr3_-_39234074 | 0.17 |
ENST00000340369.3
ENST00000421646.1 ENST00000396251.1 |
XIRP1
|
xin actin-binding repeat containing 1 |
| chr5_+_173763250 | 0.17 |
ENST00000515513.1
ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1
|
RP11-267A15.1 |
| chr16_-_47493041 | 0.17 |
ENST00000565940.2
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1 |
| chr3_-_42003479 | 0.17 |
ENST00000420927.1
|
ULK4
|
unc-51 like kinase 4 |
| chr1_+_19578033 | 0.17 |
ENST00000330263.4
|
MRTO4
|
mRNA turnover 4 homolog (S. cerevisiae) |
| chr12_-_44152551 | 0.17 |
ENST00000416848.2
ENST00000550784.1 ENST00000547156.1 ENST00000549868.1 ENST00000553166.1 ENST00000551923.1 ENST00000431332.3 ENST00000344862.5 |
PUS7L
|
pseudouridylate synthase 7 homolog (S. cerevisiae)-like |
| chr18_+_7754957 | 0.17 |
ENST00000400053.4
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr4_-_79860506 | 0.16 |
ENST00000295462.3
ENST00000380645.4 ENST00000512733.1 |
PAQR3
|
progestin and adipoQ receptor family member III |
| chr7_+_75511362 | 0.16 |
ENST00000428119.1
|
RHBDD2
|
rhomboid domain containing 2 |
| chr3_+_186435137 | 0.16 |
ENST00000447445.1
|
KNG1
|
kininogen 1 |
| chr18_-_10791648 | 0.16 |
ENST00000583325.1
|
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr8_-_30706608 | 0.16 |
ENST00000256246.2
|
TEX15
|
testis expressed 15 |
| chr3_+_32726774 | 0.16 |
ENST00000538368.1
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr6_-_72129806 | 0.15 |
ENST00000413945.1
ENST00000602878.1 ENST00000436803.1 ENST00000421704.1 ENST00000441570.1 |
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr19_+_10563567 | 0.15 |
ENST00000344979.3
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr10_-_101841588 | 0.15 |
ENST00000370418.3
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr3_+_186158169 | 0.15 |
ENST00000435548.1
ENST00000421006.1 |
RP11-78H24.1
|
RP11-78H24.1 |
| chr4_+_186317133 | 0.15 |
ENST00000507753.1
|
ANKRD37
|
ankyrin repeat domain 37 |
| chr7_+_75544466 | 0.15 |
ENST00000421059.1
ENST00000394893.1 ENST00000412521.1 ENST00000414186.1 |
POR
|
P450 (cytochrome) oxidoreductase |
| chr19_-_56110859 | 0.15 |
ENST00000221665.3
ENST00000592585.1 |
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr11_-_66360548 | 0.15 |
ENST00000333861.3
|
CCDC87
|
coiled-coil domain containing 87 |
| chr20_-_36661826 | 0.15 |
ENST00000373448.2
ENST00000373447.3 |
TTI1
|
TELO2 interacting protein 1 |
| chr2_-_69180012 | 0.15 |
ENST00000481498.1
|
GKN2
|
gastrokine 2 |
| chr20_-_21494654 | 0.15 |
ENST00000377142.4
|
NKX2-2
|
NK2 homeobox 2 |
| chr9_+_125281420 | 0.14 |
ENST00000340750.1
|
OR1J4
|
olfactory receptor, family 1, subfamily J, member 4 |
| chr9_+_4839762 | 0.14 |
ENST00000448872.2
ENST00000441844.1 |
RCL1
|
RNA terminal phosphate cyclase-like 1 |
| chr1_+_202172848 | 0.14 |
ENST00000255432.7
|
LGR6
|
leucine-rich repeat containing G protein-coupled receptor 6 |
| chr12_+_32259696 | 0.14 |
ENST00000551848.1
|
BICD1
|
bicaudal D homolog 1 (Drosophila) |
| chr22_-_24096630 | 0.14 |
ENST00000248948.3
|
VPREB3
|
pre-B lymphocyte 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.3 | 0.9 | GO:0051257 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.3 | 1.6 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.2 | 2.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 2.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.2 | 0.9 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.2 | 0.6 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.2 | 1.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.2 | 0.7 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.2 | 1.6 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 1.5 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.2 | 1.6 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.5 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.5 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.6 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 0.3 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.1 | 0.3 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.1 | 0.3 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 0.1 | 0.5 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 0.5 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 0.6 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.4 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.1 | 0.4 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 0.4 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 1.2 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.7 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.1 | 0.3 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.1 | 3.3 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.1 | 0.5 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 1.0 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.5 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 0.4 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 1.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0043602 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.4 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.1 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.5 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.7 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.4 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.5 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 0.3 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0061565 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.0 | 0.3 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.2 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.4 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.8 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.5 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 1.1 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.8 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.2 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.7 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.7 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.4 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.1 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.5 | GO:0042255 | ribosome assembly(GO:0042255) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.3 | 0.9 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.3 | 1.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 1.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 0.5 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.1 | 0.8 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 0.5 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.1 | 0.5 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 1.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.5 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 1.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.4 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.1 | 0.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 1.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.5 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.5 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 1.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.6 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.8 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 3.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.4 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 2.1 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.8 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 1.3 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.4 | 1.1 | GO:0048257 | 3'-flap endonuclease activity(GO:0048257) |
| 0.4 | 1.8 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.3 | 1.0 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.3 | 3.3 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.3 | 1.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.2 | 0.9 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.2 | 1.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 0.5 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.2 | 1.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 0.5 | GO:0048244 | phytanoyl-CoA dioxygenase activity(GO:0048244) |
| 0.1 | 0.4 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.5 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 0.7 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.9 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.1 | 0.3 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.1 | 0.3 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 0.7 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 1.0 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.3 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.1 | 1.6 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 0.5 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.5 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 0.3 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.5 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.4 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.4 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 2.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.1 | GO:0047726 | iron-cytochrome-c reductase activity(GO:0047726) |
| 0.0 | 1.5 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.6 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 1.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 2.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.2 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 1.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.6 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |