Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
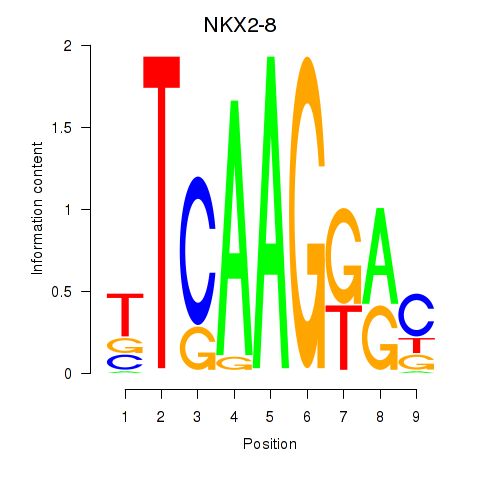
Results for NKX2-8
Z-value: 0.31
Transcription factors associated with NKX2-8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-8
|
ENSG00000136327.6 | NK2 homeobox 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-8 | hg19_v2_chr14_-_37051798_37051831 | -0.69 | 8.1e-04 | Click! |
Activity profile of NKX2-8 motif
Sorted Z-values of NKX2-8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_13349650 | 0.92 |
ENST00000256951.5
ENST00000431267.2 ENST00000542474.1 ENST00000544053.1 |
EMP1
|
epithelial membrane protein 1 |
| chr1_-_95007193 | 0.77 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr14_-_24732403 | 0.76 |
ENST00000206765.6
|
TGM1
|
transglutaminase 1 |
| chr5_-_78808617 | 0.73 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chr12_+_13349711 | 0.70 |
ENST00000538364.1
ENST00000396301.3 |
EMP1
|
epithelial membrane protein 1 |
| chr11_+_35222629 | 0.68 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr9_-_22009297 | 0.61 |
ENST00000276925.6
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr2_-_151395525 | 0.58 |
ENST00000439275.1
|
RND3
|
Rho family GTPase 3 |
| chr15_+_45422178 | 0.52 |
ENST00000389037.3
ENST00000558322.1 |
DUOX1
|
dual oxidase 1 |
| chr14_-_24732368 | 0.52 |
ENST00000544573.1
|
TGM1
|
transglutaminase 1 |
| chrX_+_115567767 | 0.49 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr8_-_29120604 | 0.48 |
ENST00000521515.1
|
KIF13B
|
kinesin family member 13B |
| chr3_-_87039662 | 0.46 |
ENST00000494229.1
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr3_+_111717511 | 0.45 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr4_-_119274121 | 0.44 |
ENST00000296498.3
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr6_+_30844192 | 0.43 |
ENST00000502955.1
ENST00000505066.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr16_+_8807419 | 0.42 |
ENST00000565016.1
ENST00000567812.1 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr1_+_151512775 | 0.41 |
ENST00000368849.3
ENST00000392712.3 ENST00000353024.3 ENST00000368848.2 ENST00000538902.1 |
TUFT1
|
tuftelin 1 |
| chr4_-_153601136 | 0.40 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr1_-_94147385 | 0.39 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr12_+_66218212 | 0.39 |
ENST00000393578.3
ENST00000425208.2 ENST00000536545.1 ENST00000354636.3 |
HMGA2
|
high mobility group AT-hook 2 |
| chr10_-_101673782 | 0.39 |
ENST00000422692.1
|
DNMBP
|
dynamin binding protein |
| chr5_+_3596168 | 0.35 |
ENST00000302006.3
|
IRX1
|
iroquois homeobox 1 |
| chr10_+_71078595 | 0.35 |
ENST00000359426.6
|
HK1
|
hexokinase 1 |
| chr1_-_31902614 | 0.34 |
ENST00000596131.1
|
AC114494.1
|
HCG1787699; Uncharacterized protein |
| chr3_+_111717600 | 0.34 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr17_+_61086917 | 0.34 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr1_+_76251912 | 0.34 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr2_-_46385 | 0.34 |
ENST00000327669.4
|
FAM110C
|
family with sequence similarity 110, member C |
| chr1_+_154401791 | 0.33 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr5_+_176692466 | 0.32 |
ENST00000508029.1
ENST00000503056.1 |
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr15_-_99057551 | 0.32 |
ENST00000558256.1
|
FAM169B
|
family with sequence similarity 169, member B |
| chr1_-_68698222 | 0.31 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr1_+_12040238 | 0.31 |
ENST00000444836.1
ENST00000235329.5 |
MFN2
|
mitofusin 2 |
| chr7_-_127672146 | 0.30 |
ENST00000476782.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr11_+_35211429 | 0.29 |
ENST00000525688.1
ENST00000278385.6 ENST00000533222.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr2_-_178129853 | 0.29 |
ENST00000397062.3
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr7_+_135242652 | 0.28 |
ENST00000285968.6
ENST00000440390.2 |
NUP205
|
nucleoporin 205kDa |
| chr4_-_100484825 | 0.28 |
ENST00000273962.3
ENST00000514547.1 ENST00000455368.2 |
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr2_-_207024233 | 0.27 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr2_+_136289030 | 0.26 |
ENST00000409478.1
ENST00000264160.4 ENST00000329971.3 ENST00000438014.1 |
R3HDM1
|
R3H domain containing 1 |
| chr22_+_21213771 | 0.26 |
ENST00000439214.1
|
SNAP29
|
synaptosomal-associated protein, 29kDa |
| chr8_+_27182862 | 0.26 |
ENST00000521164.1
ENST00000346049.5 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr11_-_10828892 | 0.25 |
ENST00000525681.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr6_-_137113604 | 0.25 |
ENST00000359015.4
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr14_-_88282595 | 0.25 |
ENST00000554519.1
|
RP11-1152H15.1
|
RP11-1152H15.1 |
| chr8_-_144679264 | 0.25 |
ENST00000531953.1
ENST00000526133.1 |
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr15_-_60683326 | 0.24 |
ENST00000559350.1
ENST00000558986.1 ENST00000560389.1 |
ANXA2
|
annexin A2 |
| chr2_-_207023918 | 0.24 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr3_+_63953415 | 0.24 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr9_-_128412696 | 0.24 |
ENST00000420643.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr17_-_8055747 | 0.24 |
ENST00000317276.4
ENST00000581703.1 |
PER1
|
period circadian clock 1 |
| chr12_+_66218903 | 0.23 |
ENST00000393577.3
|
HMGA2
|
high mobility group AT-hook 2 |
| chr18_-_72920372 | 0.23 |
ENST00000581620.1
ENST00000582437.1 |
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr20_+_58296265 | 0.23 |
ENST00000395636.2
ENST00000361300.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr1_-_115292591 | 0.22 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr8_+_104310661 | 0.22 |
ENST00000522566.1
|
FZD6
|
frizzled family receptor 6 |
| chr3_+_187930429 | 0.22 |
ENST00000420410.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr9_+_109685630 | 0.22 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr11_-_128457446 | 0.22 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr12_+_25205568 | 0.22 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr1_+_152957707 | 0.22 |
ENST00000368762.1
|
SPRR1A
|
small proline-rich protein 1A |
| chr13_+_28712614 | 0.22 |
ENST00000380958.3
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr2_-_207024134 | 0.21 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr6_-_32145861 | 0.21 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr4_-_7873713 | 0.21 |
ENST00000382543.3
|
AFAP1
|
actin filament associated protein 1 |
| chr21_-_35899113 | 0.21 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr1_+_16083098 | 0.20 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chrX_+_123094672 | 0.20 |
ENST00000354548.5
ENST00000458700.1 |
STAG2
|
stromal antigen 2 |
| chr1_+_55464600 | 0.19 |
ENST00000371265.4
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr3_-_16524357 | 0.19 |
ENST00000432519.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr18_-_44497308 | 0.19 |
ENST00000585916.1
ENST00000324794.7 ENST00000545673.1 |
PIAS2
|
protein inhibitor of activated STAT, 2 |
| chr4_+_144303093 | 0.19 |
ENST00000505913.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr5_+_43602750 | 0.19 |
ENST00000505678.2
ENST00000512422.1 ENST00000264663.5 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr6_+_143772060 | 0.19 |
ENST00000367591.4
|
PEX3
|
peroxisomal biogenesis factor 3 |
| chr14_-_36990354 | 0.19 |
ENST00000518149.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr10_+_123951957 | 0.19 |
ENST00000514539.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr6_-_131299929 | 0.18 |
ENST00000531356.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr16_+_23194033 | 0.18 |
ENST00000300061.2
|
SCNN1G
|
sodium channel, non-voltage-gated 1, gamma subunit |
| chr12_+_25205666 | 0.18 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr12_+_25205446 | 0.18 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr15_+_74911430 | 0.18 |
ENST00000562670.1
ENST00000564096.1 |
CLK3
|
CDC-like kinase 3 |
| chr6_+_158957431 | 0.18 |
ENST00000367090.3
|
TMEM181
|
transmembrane protein 181 |
| chr11_-_117170403 | 0.18 |
ENST00000504995.1
|
BACE1
|
beta-site APP-cleaving enzyme 1 |
| chr4_-_100485183 | 0.18 |
ENST00000394876.2
|
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr9_+_131873842 | 0.18 |
ENST00000417728.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr17_-_62503015 | 0.18 |
ENST00000581806.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chrX_-_64196376 | 0.18 |
ENST00000447788.2
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr13_-_23949671 | 0.18 |
ENST00000402364.1
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr20_-_57089934 | 0.18 |
ENST00000439429.1
ENST00000371149.3 |
APCDD1L
|
adenomatosis polyposis coli down-regulated 1-like |
| chrX_-_64196351 | 0.17 |
ENST00000374839.3
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr14_+_76618242 | 0.17 |
ENST00000557542.1
ENST00000557263.1 ENST00000557207.1 ENST00000312858.5 ENST00000261530.7 |
GPATCH2L
|
G patch domain containing 2-like |
| chr3_-_187455680 | 0.17 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr14_+_58894404 | 0.17 |
ENST00000554463.1
ENST00000555833.1 |
KIAA0586
|
KIAA0586 |
| chr12_+_121837905 | 0.17 |
ENST00000392465.3
ENST00000554606.1 ENST00000392464.2 ENST00000555076.1 |
RNF34
|
ring finger protein 34, E3 ubiquitin protein ligase |
| chrX_-_32173579 | 0.17 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr11_+_27062272 | 0.17 |
ENST00000529202.1
ENST00000533566.1 |
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr8_+_107593198 | 0.17 |
ENST00000517686.1
|
OXR1
|
oxidation resistance 1 |
| chr17_-_46178527 | 0.17 |
ENST00000393408.3
|
CBX1
|
chromobox homolog 1 |
| chr12_-_46663734 | 0.17 |
ENST00000550173.1
|
SLC38A1
|
solute carrier family 38, member 1 |
| chr22_+_20877924 | 0.17 |
ENST00000445189.1
|
MED15
|
mediator complex subunit 15 |
| chr7_+_18330035 | 0.16 |
ENST00000413509.2
|
HDAC9
|
histone deacetylase 9 |
| chr13_-_46756351 | 0.16 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr2_-_110371720 | 0.16 |
ENST00000356688.4
|
SEPT10
|
septin 10 |
| chr5_+_14664762 | 0.16 |
ENST00000284274.4
|
FAM105B
|
family with sequence similarity 105, member B |
| chr14_+_37667230 | 0.15 |
ENST00000556451.1
ENST00000556753.1 ENST00000396294.2 |
MIPOL1
|
mirror-image polydactyly 1 |
| chr7_-_137686791 | 0.15 |
ENST00000452463.1
ENST00000330387.6 ENST00000456390.1 |
CREB3L2
|
cAMP responsive element binding protein 3-like 2 |
| chr5_+_34929677 | 0.15 |
ENST00000342382.4
ENST00000382021.2 ENST00000303525.7 |
DNAJC21
|
DnaJ (Hsp40) homolog, subfamily C, member 21 |
| chr22_-_30925150 | 0.15 |
ENST00000437871.1
|
SEC14L6
|
SEC14-like 6 (S. cerevisiae) |
| chr3_-_52739670 | 0.15 |
ENST00000497953.1
|
GLT8D1
|
glycosyltransferase 8 domain containing 1 |
| chr2_+_119699864 | 0.15 |
ENST00000541757.1
ENST00000412481.1 |
MARCO
|
macrophage receptor with collagenous structure |
| chr20_+_55967129 | 0.15 |
ENST00000371219.2
|
RBM38
|
RNA binding motif protein 38 |
| chr14_-_107219365 | 0.14 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr3_-_125094093 | 0.14 |
ENST00000484491.1
ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148
|
zinc finger protein 148 |
| chr10_+_24755416 | 0.14 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr17_+_34901353 | 0.14 |
ENST00000593016.1
|
GGNBP2
|
gametogenetin binding protein 2 |
| chr12_+_25205155 | 0.14 |
ENST00000550945.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr7_+_30185496 | 0.14 |
ENST00000455738.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chrX_-_64196307 | 0.14 |
ENST00000545618.1
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chrX_+_118533269 | 0.13 |
ENST00000336249.7
|
SLC25A43
|
solute carrier family 25, member 43 |
| chr11_-_77348796 | 0.13 |
ENST00000263309.3
ENST00000525064.1 |
CLNS1A
|
chloride channel, nucleotide-sensitive, 1A |
| chr1_+_109289279 | 0.13 |
ENST00000370008.3
|
STXBP3
|
syntaxin binding protein 3 |
| chr22_+_29834572 | 0.13 |
ENST00000354373.2
|
RFPL1
|
ret finger protein-like 1 |
| chr12_-_95397442 | 0.13 |
ENST00000547157.1
ENST00000547986.1 ENST00000327772.2 |
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr1_+_10057274 | 0.13 |
ENST00000294435.7
|
RBP7
|
retinol binding protein 7, cellular |
| chr20_-_48747662 | 0.13 |
ENST00000371656.2
|
TMEM189
|
transmembrane protein 189 |
| chr8_-_141810634 | 0.13 |
ENST00000521986.1
ENST00000523539.1 ENST00000538769.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr4_+_141264597 | 0.13 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr1_+_224301787 | 0.12 |
ENST00000366862.5
ENST00000424254.2 |
FBXO28
|
F-box protein 28 |
| chr8_-_145018905 | 0.12 |
ENST00000398774.2
|
PLEC
|
plectin |
| chr3_+_187930491 | 0.12 |
ENST00000443217.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr14_+_58765103 | 0.12 |
ENST00000355431.3
ENST00000348476.3 ENST00000395168.3 |
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr17_+_46185111 | 0.11 |
ENST00000582104.1
ENST00000584335.1 |
SNX11
|
sorting nexin 11 |
| chr7_-_86974767 | 0.11 |
ENST00000610086.1
|
TP53TG1
|
TP53 target 1 (non-protein coding) |
| chr1_+_16083123 | 0.11 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr5_+_68665608 | 0.11 |
ENST00000509734.1
ENST00000354868.5 ENST00000521422.1 ENST00000354312.3 ENST00000345306.6 |
RAD17
|
RAD17 homolog (S. pombe) |
| chr2_+_119699742 | 0.11 |
ENST00000327097.4
|
MARCO
|
macrophage receptor with collagenous structure |
| chr12_+_60058458 | 0.10 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr7_+_30634297 | 0.10 |
ENST00000389266.3
|
GARS
|
glycyl-tRNA synthetase |
| chr7_+_112063192 | 0.10 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chrX_+_123094369 | 0.10 |
ENST00000455404.1
ENST00000218089.9 |
STAG2
|
stromal antigen 2 |
| chr22_+_38609538 | 0.10 |
ENST00000407965.1
|
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr7_-_127671674 | 0.10 |
ENST00000478726.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr17_+_71228793 | 0.09 |
ENST00000426147.2
|
C17orf80
|
chromosome 17 open reading frame 80 |
| chr4_-_100485143 | 0.09 |
ENST00000394877.3
|
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr11_-_104827425 | 0.09 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr2_-_106054952 | 0.09 |
ENST00000336660.5
ENST00000393352.3 ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr1_+_173793777 | 0.09 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr3_+_39093481 | 0.09 |
ENST00000302313.5
ENST00000544962.1 ENST00000396258.3 ENST00000418020.1 |
WDR48
|
WD repeat domain 48 |
| chr3_+_4535025 | 0.09 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr2_+_62132781 | 0.08 |
ENST00000311832.5
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr16_-_67493110 | 0.08 |
ENST00000602876.1
|
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr3_+_126423045 | 0.08 |
ENST00000290913.3
ENST00000508789.1 |
CHCHD6
|
coiled-coil-helix-coiled-coil-helix domain containing 6 |
| chr2_-_197675000 | 0.08 |
ENST00000342506.2
|
C2orf66
|
chromosome 2 open reading frame 66 |
| chr11_-_19223523 | 0.08 |
ENST00000265968.3
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr3_+_177545563 | 0.08 |
ENST00000434309.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr14_-_78083112 | 0.08 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr14_+_58765305 | 0.08 |
ENST00000445108.1
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr1_-_104238912 | 0.08 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr19_+_52074502 | 0.08 |
ENST00000545217.1
ENST00000262259.2 ENST00000596504.1 |
ZNF175
|
zinc finger protein 175 |
| chr17_-_62502639 | 0.08 |
ENST00000225792.5
ENST00000581697.1 ENST00000584279.1 ENST00000577922.1 |
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr9_-_99637820 | 0.08 |
ENST00000289032.8
ENST00000535338.1 |
ZNF782
|
zinc finger protein 782 |
| chr22_+_39101728 | 0.08 |
ENST00000216044.5
ENST00000484657.1 |
GTPBP1
|
GTP binding protein 1 |
| chr11_-_77348821 | 0.08 |
ENST00000528364.1
ENST00000532069.1 ENST00000525428.1 |
CLNS1A
|
chloride channel, nucleotide-sensitive, 1A |
| chr3_-_47950745 | 0.07 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr11_+_59705928 | 0.07 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr14_-_106068065 | 0.07 |
ENST00000390541.2
|
IGHE
|
immunoglobulin heavy constant epsilon |
| chr19_-_44259053 | 0.07 |
ENST00000601170.1
|
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr19_-_11347173 | 0.07 |
ENST00000587656.1
|
DOCK6
|
dedicator of cytokinesis 6 |
| chrM_+_8366 | 0.07 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr2_-_136288740 | 0.06 |
ENST00000264159.6
ENST00000536680.1 |
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr3_+_10183447 | 0.06 |
ENST00000345392.2
|
VHL
|
von Hippel-Lindau tumor suppressor, E3 ubiquitin protein ligase |
| chrX_+_72064690 | 0.06 |
ENST00000438696.1
|
DMRTC1B
|
DMRT-like family C1B |
| chr16_-_3350614 | 0.06 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chrM_+_8527 | 0.06 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chr12_+_66218598 | 0.06 |
ENST00000541363.1
|
HMGA2
|
high mobility group AT-hook 2 |
| chr12_+_25205628 | 0.06 |
ENST00000554942.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr20_+_42143136 | 0.06 |
ENST00000373134.1
|
L3MBTL1
|
l(3)mbt-like 1 (Drosophila) |
| chr19_-_17622269 | 0.06 |
ENST00000595116.1
|
CTD-3131K8.2
|
CTD-3131K8.2 |
| chr11_+_925824 | 0.06 |
ENST00000525796.1
ENST00000534328.1 ENST00000448903.2 ENST00000332231.5 |
AP2A2
|
adaptor-related protein complex 2, alpha 2 subunit |
| chr6_-_18249971 | 0.05 |
ENST00000507591.1
|
DEK
|
DEK oncogene |
| chr11_-_10715117 | 0.05 |
ENST00000527509.2
|
MRVI1
|
murine retrovirus integration site 1 homolog |
| chr18_-_24722995 | 0.05 |
ENST00000581714.1
|
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr14_-_106518922 | 0.05 |
ENST00000390598.2
|
IGHV3-7
|
immunoglobulin heavy variable 3-7 |
| chr14_-_107199464 | 0.05 |
ENST00000433072.2
|
IGHV3-72
|
immunoglobulin heavy variable 3-72 |
| chr15_-_70387120 | 0.05 |
ENST00000539550.1
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr17_+_39994032 | 0.05 |
ENST00000293303.4
ENST00000438813.1 |
KLHL10
|
kelch-like family member 10 |
| chr3_+_101498026 | 0.05 |
ENST00000495842.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr2_+_207024306 | 0.05 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr3_-_53164423 | 0.05 |
ENST00000467048.1
ENST00000394738.3 ENST00000296292.3 |
RFT1
|
RFT1 homolog (S. cerevisiae) |
| chr19_-_3600549 | 0.05 |
ENST00000589966.1
|
TBXA2R
|
thromboxane A2 receptor |
| chr17_+_62503073 | 0.05 |
ENST00000580188.1
ENST00000581056.1 |
CEP95
|
centrosomal protein 95kDa |
| chr14_-_106791536 | 0.05 |
ENST00000390613.2
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr22_+_32753854 | 0.04 |
ENST00000249007.4
|
RFPL3
|
ret finger protein-like 3 |
| chr7_+_73507409 | 0.04 |
ENST00000538333.3
|
LIMK1
|
LIM domain kinase 1 |
| chr2_-_55277436 | 0.04 |
ENST00000354474.6
|
RTN4
|
reticulon 4 |
| chr14_+_37667193 | 0.04 |
ENST00000539062.2
|
MIPOL1
|
mirror-image polydactyly 1 |
| chr22_+_38453207 | 0.04 |
ENST00000404072.3
ENST00000424694.1 |
PICK1
|
protein interacting with PRKCA 1 |
| chr10_-_135171178 | 0.04 |
ENST00000368551.1
|
FUOM
|
fucose mutarotase |
| chr16_+_4606347 | 0.04 |
ENST00000444310.4
|
C16orf96
|
chromosome 16 open reading frame 96 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.2 | 1.3 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.2 | 0.7 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.2 | 0.7 | GO:0003131 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.1 | 1.0 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.4 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 0.5 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.4 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.1 | 0.3 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 0.3 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.2 | GO:0052361 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.1 | 0.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.2 | GO:2000374 | regulation of oxygen metabolic process(GO:2000374) |
| 0.1 | 0.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 1.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.3 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.2 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.3 | GO:0046223 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.5 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.0 | 0.3 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.4 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.2 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.1 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.2 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.3 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.3 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.1 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.3 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.1 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.5 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.4 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.7 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.1 | 0.4 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 0.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.7 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.2 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.2 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.3 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.8 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 1.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.1 | 0.3 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.1 | 0.6 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 1.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.4 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 0.4 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.1 | 0.2 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.3 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.3 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.4 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.1 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.4 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.5 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.8 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |