Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
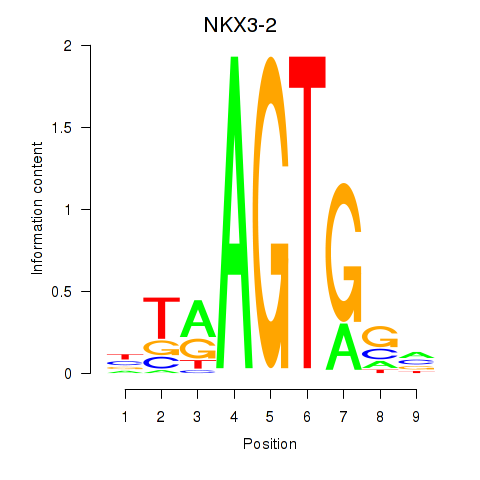
Results for NKX3-2
Z-value: 0.64
Transcription factors associated with NKX3-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX3-2
|
ENSG00000109705.7 | NK3 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX3-2 | hg19_v2_chr4_-_13546632_13546674 | -0.24 | 3.1e-01 | Click! |
Activity profile of NKX3-2 motif
Sorted Z-values of NKX3-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_193853927 | 0.79 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr1_+_205473720 | 0.73 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr12_+_4382917 | 0.68 |
ENST00000261254.3
|
CCND2
|
cyclin D2 |
| chr11_-_119993979 | 0.63 |
ENST00000524816.3
ENST00000525327.1 |
TRIM29
|
tripartite motif containing 29 |
| chr2_-_25451065 | 0.62 |
ENST00000606328.1
|
RP11-458N5.1
|
RP11-458N5.1 |
| chr10_-_98031265 | 0.62 |
ENST00000224337.5
ENST00000371176.2 |
BLNK
|
B-cell linker |
| chr19_-_6720686 | 0.62 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr19_-_6670128 | 0.60 |
ENST00000245912.3
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr10_-_98031310 | 0.60 |
ENST00000427367.2
ENST00000413476.2 |
BLNK
|
B-cell linker |
| chr12_-_10022735 | 0.59 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr15_-_31521567 | 0.55 |
ENST00000560812.1
ENST00000559853.1 ENST00000558109.1 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr10_-_95360983 | 0.53 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr19_+_18544045 | 0.53 |
ENST00000599699.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr10_-_7708918 | 0.52 |
ENST00000256861.6
ENST00000397146.2 ENST00000446830.2 ENST00000397145.2 |
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr2_+_17721920 | 0.51 |
ENST00000295156.4
|
VSNL1
|
visinin-like 1 |
| chr1_+_26606608 | 0.51 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr17_-_1619568 | 0.49 |
ENST00000571595.1
|
MIR22HG
|
MIR22 host gene (non-protein coding) |
| chr19_+_7694623 | 0.48 |
ENST00000594797.1
ENST00000456958.3 ENST00000601406.1 |
PET100
|
PET100 homolog (S. cerevisiae) |
| chr11_-_59633951 | 0.45 |
ENST00000257264.3
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr6_-_32784687 | 0.44 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr19_-_36297348 | 0.44 |
ENST00000589835.1
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr1_+_205473784 | 0.44 |
ENST00000478560.1
ENST00000443813.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr11_+_35222629 | 0.43 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr20_-_39995467 | 0.42 |
ENST00000332312.3
|
EMILIN3
|
elastin microfibril interfacer 3 |
| chr8_-_49833978 | 0.42 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr2_+_208104497 | 0.41 |
ENST00000430494.1
|
AC007879.7
|
AC007879.7 |
| chr1_+_152974218 | 0.41 |
ENST00000331860.3
ENST00000443178.1 ENST00000295367.4 |
SPRR3
|
small proline-rich protein 3 |
| chr19_+_48867652 | 0.40 |
ENST00000344846.2
|
SYNGR4
|
synaptogyrin 4 |
| chr15_+_41136734 | 0.40 |
ENST00000568580.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr3_+_50192537 | 0.40 |
ENST00000002829.3
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr3_+_50192499 | 0.40 |
ENST00000413852.1
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chrX_+_105937068 | 0.39 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr19_+_54609230 | 0.39 |
ENST00000420296.1
|
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
| chr11_+_64879317 | 0.39 |
ENST00000526809.1
ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2
|
transmembrane 7 superfamily member 2 |
| chr1_+_86889769 | 0.39 |
ENST00000370565.4
|
CLCA2
|
chloride channel accessory 2 |
| chr7_-_45957011 | 0.39 |
ENST00000417621.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr12_-_50616122 | 0.39 |
ENST00000552823.1
ENST00000552909.1 |
LIMA1
|
LIM domain and actin binding 1 |
| chr8_-_49834299 | 0.38 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr2_-_25016251 | 0.38 |
ENST00000328379.5
|
PTRHD1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chrX_+_105936982 | 0.38 |
ENST00000418562.1
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr9_-_22009297 | 0.36 |
ENST00000276925.6
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr5_-_149792295 | 0.36 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr22_+_38453207 | 0.36 |
ENST00000404072.3
ENST00000424694.1 |
PICK1
|
protein interacting with PRKCA 1 |
| chr12_+_56473939 | 0.34 |
ENST00000450146.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr22_-_31328881 | 0.34 |
ENST00000445980.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr19_-_5680202 | 0.34 |
ENST00000590389.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr1_-_204183071 | 0.34 |
ENST00000308302.3
|
GOLT1A
|
golgi transport 1A |
| chr14_+_78227105 | 0.33 |
ENST00000439131.2
ENST00000355883.3 ENST00000557011.1 ENST00000556047.1 |
C14orf178
|
chromosome 14 open reading frame 178 |
| chr8_-_29592736 | 0.33 |
ENST00000518623.1
|
LINC00589
|
long intergenic non-protein coding RNA 589 |
| chr3_-_107777208 | 0.33 |
ENST00000398258.3
|
CD47
|
CD47 molecule |
| chr19_-_10444188 | 0.33 |
ENST00000293677.6
|
RAVER1
|
ribonucleoprotein, PTB-binding 1 |
| chr7_-_45956856 | 0.33 |
ENST00000428530.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr10_-_131762105 | 0.32 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr1_+_149822620 | 0.32 |
ENST00000369159.2
|
HIST2H2AA4
|
histone cluster 2, H2aa4 |
| chr5_-_77072085 | 0.32 |
ENST00000518338.2
ENST00000520039.1 ENST00000306388.6 ENST00000520361.1 |
TBCA
|
tubulin folding cofactor A |
| chr9_+_35906176 | 0.32 |
ENST00000354323.2
|
HRCT1
|
histidine rich carboxyl terminus 1 |
| chr7_-_102213030 | 0.31 |
ENST00000511313.1
ENST00000513438.1 ENST00000513506.1 |
POLR2J3
|
polymerase (RNA) II (DNA directed) polypeptide J3 |
| chr10_+_7745232 | 0.31 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr7_+_90339169 | 0.31 |
ENST00000436577.2
|
CDK14
|
cyclin-dependent kinase 14 |
| chr11_+_128563652 | 0.31 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr14_+_21566980 | 0.31 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chrX_+_51928002 | 0.31 |
ENST00000375626.3
|
MAGED4
|
melanoma antigen family D, 4 |
| chr8_+_35649365 | 0.31 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr9_+_34458771 | 0.31 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr20_-_2821271 | 0.30 |
ENST00000448755.1
ENST00000360652.2 |
PCED1A
|
PC-esterase domain containing 1A |
| chr11_+_66045634 | 0.30 |
ENST00000528852.1
ENST00000311445.6 |
CNIH2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr7_-_8276508 | 0.30 |
ENST00000401396.1
ENST00000317367.5 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr3_-_69249863 | 0.30 |
ENST00000478263.1
ENST00000462512.1 |
FRMD4B
|
FERM domain containing 4B |
| chrX_+_48660287 | 0.29 |
ENST00000444343.2
ENST00000376610.2 ENST00000334136.5 ENST00000376619.2 |
HDAC6
|
histone deacetylase 6 |
| chr22_-_21356375 | 0.29 |
ENST00000215742.4
ENST00000399133.2 |
THAP7
|
THAP domain containing 7 |
| chr14_-_62162541 | 0.29 |
ENST00000557544.1
|
HIF1A-AS1
|
HIF1A antisense RNA 1 |
| chr21_-_33985127 | 0.29 |
ENST00000290155.3
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr20_+_1115821 | 0.29 |
ENST00000435720.1
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr1_-_149814478 | 0.29 |
ENST00000369161.3
|
HIST2H2AA3
|
histone cluster 2, H2aa3 |
| chr1_-_155145721 | 0.28 |
ENST00000295682.4
|
KRTCAP2
|
keratinocyte associated protein 2 |
| chr7_+_90338712 | 0.28 |
ENST00000265741.3
ENST00000406263.1 |
CDK14
|
cyclin-dependent kinase 14 |
| chr12_-_113658892 | 0.28 |
ENST00000299732.2
ENST00000416617.2 |
IQCD
|
IQ motif containing D |
| chr2_-_27294500 | 0.28 |
ENST00000447619.1
ENST00000429985.1 ENST00000456793.1 |
OST4
|
oligosaccharyltransferase 4 homolog (S. cerevisiae) |
| chr19_-_36545649 | 0.28 |
ENST00000292894.1
|
THAP8
|
THAP domain containing 8 |
| chr21_-_33984888 | 0.28 |
ENST00000382549.4
ENST00000540881.1 |
C21orf59
|
chromosome 21 open reading frame 59 |
| chr12_+_6494285 | 0.27 |
ENST00000541102.1
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr10_-_47173994 | 0.27 |
ENST00000414655.2
ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1
LINC00842
|
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr14_+_21156915 | 0.27 |
ENST00000397990.4
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr1_-_153013588 | 0.27 |
ENST00000360379.3
|
SPRR2D
|
small proline-rich protein 2D |
| chr2_-_42160486 | 0.27 |
ENST00000427054.1
|
AC104654.2
|
AC104654.2 |
| chr17_+_68071389 | 0.27 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr10_+_7745303 | 0.26 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr1_+_149230680 | 0.26 |
ENST00000443018.1
|
RP11-403I13.5
|
RP11-403I13.5 |
| chr8_+_145162629 | 0.26 |
ENST00000323662.8
|
KIAA1875
|
KIAA1875 |
| chr16_+_30671223 | 0.26 |
ENST00000568722.1
|
FBRS
|
fibrosin |
| chr3_-_42003613 | 0.26 |
ENST00000414606.1
|
ULK4
|
unc-51 like kinase 4 |
| chr9_-_22009241 | 0.26 |
ENST00000380142.4
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr6_+_29910301 | 0.26 |
ENST00000376809.5
ENST00000376802.2 |
HLA-A
|
major histocompatibility complex, class I, A |
| chr3_-_128712955 | 0.26 |
ENST00000265068.5
|
KIAA1257
|
KIAA1257 |
| chr17_+_7482785 | 0.25 |
ENST00000250092.6
ENST00000380498.6 ENST00000584502.1 |
CD68
|
CD68 molecule |
| chr15_+_42694573 | 0.25 |
ENST00000397200.4
ENST00000569827.1 |
CAPN3
|
calpain 3, (p94) |
| chr19_-_5680231 | 0.25 |
ENST00000587950.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr8_+_22414182 | 0.24 |
ENST00000524057.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chrX_+_153665248 | 0.24 |
ENST00000447750.2
|
GDI1
|
GDP dissociation inhibitor 1 |
| chr19_+_843314 | 0.24 |
ENST00000544537.2
|
PRTN3
|
proteinase 3 |
| chr11_-_5537920 | 0.24 |
ENST00000380184.1
|
UBQLNL
|
ubiquilin-like |
| chr7_-_99573677 | 0.24 |
ENST00000292401.4
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr11_-_119993734 | 0.24 |
ENST00000533302.1
|
TRIM29
|
tripartite motif containing 29 |
| chr17_+_34171081 | 0.24 |
ENST00000585577.1
|
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr7_-_102119342 | 0.23 |
ENST00000393794.3
ENST00000292614.5 |
POLR2J
|
polymerase (RNA) II (DNA directed) polypeptide J, 13.3kDa |
| chr2_-_182545603 | 0.23 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr7_-_135412925 | 0.23 |
ENST00000354042.4
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr7_+_30174668 | 0.23 |
ENST00000415604.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr12_-_95397442 | 0.23 |
ENST00000547157.1
ENST00000547986.1 ENST00000327772.2 |
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chrX_+_44732757 | 0.23 |
ENST00000377967.4
ENST00000536777.1 ENST00000382899.4 ENST00000543216.1 |
KDM6A
|
lysine (K)-specific demethylase 6A |
| chr12_-_50616382 | 0.23 |
ENST00000552783.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr1_+_6845497 | 0.23 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr6_-_47010061 | 0.23 |
ENST00000371253.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr2_+_220379052 | 0.23 |
ENST00000347842.3
ENST00000358078.4 |
ASIC4
|
acid-sensing (proton-gated) ion channel family member 4 |
| chr17_+_18380051 | 0.23 |
ENST00000581545.1
ENST00000582333.1 ENST00000328114.6 ENST00000412421.2 ENST00000583322.1 ENST00000584941.1 |
LGALS9C
|
lectin, galactoside-binding, soluble, 9C |
| chr11_+_67250490 | 0.22 |
ENST00000528641.2
ENST00000279146.3 |
AIP
|
aryl hydrocarbon receptor interacting protein |
| chr11_-_64511789 | 0.22 |
ENST00000419843.1
ENST00000394430.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr5_-_127418573 | 0.22 |
ENST00000508353.1
ENST00000508878.1 ENST00000501652.1 ENST00000514409.1 |
CTC-228N24.3
|
CTC-228N24.3 |
| chr20_-_2821756 | 0.22 |
ENST00000356872.3
ENST00000439542.1 |
PCED1A
|
PC-esterase domain containing 1A |
| chr8_+_87111059 | 0.22 |
ENST00000285393.3
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr15_-_74726283 | 0.22 |
ENST00000543145.2
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chr15_+_75639951 | 0.22 |
ENST00000564784.1
ENST00000569035.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr17_+_76126842 | 0.21 |
ENST00000590426.1
ENST00000590799.1 ENST00000318430.5 ENST00000589691.1 |
TMC8
|
transmembrane channel-like 8 |
| chr1_+_6845578 | 0.21 |
ENST00000467404.2
ENST00000439411.2 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr3_-_52008016 | 0.21 |
ENST00000489595.2
ENST00000461108.1 ENST00000395008.2 ENST00000525795.1 ENST00000361143.5 |
RP11-155D18.14
ABHD14B
|
Poly(rC)-binding protein 4 abhydrolase domain containing 14B |
| chr9_-_74979420 | 0.21 |
ENST00000343431.2
ENST00000376956.3 |
ZFAND5
|
zinc finger, AN1-type domain 5 |
| chrX_+_13707235 | 0.21 |
ENST00000464506.1
|
RAB9A
|
RAB9A, member RAS oncogene family |
| chr19_+_41497178 | 0.21 |
ENST00000324071.4
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chrX_-_15332665 | 0.21 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr1_+_202183200 | 0.21 |
ENST00000439764.2
|
LGR6
|
leucine-rich repeat containing G protein-coupled receptor 6 |
| chr3_+_48481658 | 0.20 |
ENST00000438607.2
|
TMA7
|
translation machinery associated 7 homolog (S. cerevisiae) |
| chr17_+_45810594 | 0.20 |
ENST00000177694.1
|
TBX21
|
T-box 21 |
| chr6_+_32821924 | 0.20 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr8_-_101734170 | 0.20 |
ENST00000522387.1
ENST00000518196.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr21_-_33984865 | 0.20 |
ENST00000458138.1
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr15_+_81475047 | 0.20 |
ENST00000559388.1
|
IL16
|
interleukin 16 |
| chr15_-_40401062 | 0.20 |
ENST00000354670.4
ENST00000559701.1 ENST00000557870.1 ENST00000558774.1 |
BMF
|
Bcl2 modifying factor |
| chr6_-_47009996 | 0.19 |
ENST00000371243.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr9_-_34458531 | 0.19 |
ENST00000379089.1
ENST00000379087.1 ENST00000379084.1 ENST00000379081.1 ENST00000379080.1 ENST00000422409.1 ENST00000379078.1 ENST00000445726.1 ENST00000297620.4 |
FAM219A
|
family with sequence similarity 219, member A |
| chr9_+_109694914 | 0.19 |
ENST00000542028.1
|
ZNF462
|
zinc finger protein 462 |
| chr7_-_150780487 | 0.19 |
ENST00000482202.1
|
TMUB1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr8_-_90769422 | 0.19 |
ENST00000524190.1
ENST00000523859.1 |
RP11-37B2.1
|
RP11-37B2.1 |
| chr19_+_16435625 | 0.19 |
ENST00000248071.5
ENST00000592003.1 |
KLF2
|
Kruppel-like factor 2 |
| chr10_+_23384435 | 0.19 |
ENST00000376510.3
|
MSRB2
|
methionine sulfoxide reductase B2 |
| chr7_-_22862406 | 0.19 |
ENST00000372879.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr11_-_93474645 | 0.19 |
ENST00000532455.1
|
TAF1D
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, D, 41kDa |
| chr20_+_32399093 | 0.18 |
ENST00000217402.2
|
CHMP4B
|
charged multivesicular body protein 4B |
| chr12_-_54978086 | 0.18 |
ENST00000553113.1
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr1_-_35497506 | 0.18 |
ENST00000317538.5
ENST00000373340.2 ENST00000357182.4 |
ZMYM6
|
zinc finger, MYM-type 6 |
| chr1_+_116519112 | 0.18 |
ENST00000369503.4
|
SLC22A15
|
solute carrier family 22, member 15 |
| chr7_+_29234375 | 0.18 |
ENST00000409350.1
ENST00000495789.2 ENST00000539389.1 |
CHN2
|
chimerin 2 |
| chr12_-_21927736 | 0.18 |
ENST00000240662.2
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr7_+_99971129 | 0.18 |
ENST00000394000.2
ENST00000350573.2 |
PILRA
|
paired immunoglobin-like type 2 receptor alpha |
| chrX_-_73051037 | 0.18 |
ENST00000445814.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr1_+_155146318 | 0.18 |
ENST00000368385.4
ENST00000545012.1 ENST00000392451.2 ENST00000368383.3 ENST00000368382.1 ENST00000334634.4 |
TRIM46
|
tripartite motif containing 46 |
| chr17_+_38083977 | 0.18 |
ENST00000578802.1
ENST00000578478.1 ENST00000582263.1 |
RP11-387H17.4
|
RP11-387H17.4 |
| chr1_+_35247859 | 0.18 |
ENST00000373362.3
|
GJB3
|
gap junction protein, beta 3, 31kDa |
| chr12_-_21928515 | 0.18 |
ENST00000537950.1
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr4_-_52883786 | 0.18 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr9_-_23821842 | 0.17 |
ENST00000544538.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr6_+_37012607 | 0.17 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr3_+_50192457 | 0.17 |
ENST00000414301.1
ENST00000450338.1 |
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chrX_-_49965663 | 0.17 |
ENST00000376056.2
ENST00000376058.2 ENST00000358526.2 |
AKAP4
|
A kinase (PRKA) anchor protein 4 |
| chr17_+_43238438 | 0.17 |
ENST00000593138.1
ENST00000586681.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr3_+_177159695 | 0.17 |
ENST00000442937.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr22_+_38453378 | 0.17 |
ENST00000437453.1
ENST00000356976.3 |
PICK1
|
protein interacting with PRKCA 1 |
| chr20_+_2821340 | 0.17 |
ENST00000380445.3
ENST00000380469.3 |
VPS16
|
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr3_-_100551141 | 0.17 |
ENST00000478235.1
ENST00000471901.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr7_-_120498357 | 0.17 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr19_+_36266417 | 0.17 |
ENST00000378944.5
ENST00000007510.4 |
ARHGAP33
|
Rho GTPase activating protein 33 |
| chrY_+_15418467 | 0.17 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr7_-_128415844 | 0.17 |
ENST00000249389.2
|
OPN1SW
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr5_+_140810132 | 0.17 |
ENST00000252085.3
|
PCDHGA12
|
protocadherin gamma subfamily A, 12 |
| chr6_+_42928485 | 0.17 |
ENST00000372808.3
|
GNMT
|
glycine N-methyltransferase |
| chr19_+_16999654 | 0.17 |
ENST00000248076.3
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr18_+_28898052 | 0.17 |
ENST00000257192.4
|
DSG1
|
desmoglein 1 |
| chr3_+_48507210 | 0.16 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr11_-_118047376 | 0.16 |
ENST00000278947.5
|
SCN2B
|
sodium channel, voltage-gated, type II, beta subunit |
| chr17_-_74137374 | 0.16 |
ENST00000322957.6
|
FOXJ1
|
forkhead box J1 |
| chr15_-_85259294 | 0.16 |
ENST00000558217.1
ENST00000558196.1 ENST00000558134.1 |
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr1_+_211433275 | 0.16 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr9_+_5890802 | 0.16 |
ENST00000381477.3
ENST00000381476.1 ENST00000381471.1 |
MLANA
|
melan-A |
| chr7_+_129007964 | 0.16 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chrX_-_103087136 | 0.16 |
ENST00000243298.2
|
RAB9B
|
RAB9B, member RAS oncogene family |
| chr15_+_90895471 | 0.16 |
ENST00000354377.3
ENST00000379090.5 |
ZNF774
|
zinc finger protein 774 |
| chr4_+_156680518 | 0.16 |
ENST00000513437.1
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr17_+_68071458 | 0.16 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr15_+_89402148 | 0.16 |
ENST00000560601.1
|
ACAN
|
aggrecan |
| chr19_+_17337473 | 0.16 |
ENST00000598068.1
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr6_-_30080876 | 0.16 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr3_-_50541028 | 0.16 |
ENST00000266039.3
ENST00000435965.1 ENST00000395083.1 |
CACNA2D2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2 |
| chr1_+_210111534 | 0.15 |
ENST00000422431.1
ENST00000534859.1 ENST00000399639.2 ENST00000537238.1 |
SYT14
|
synaptotagmin XIV |
| chr17_-_46507537 | 0.15 |
ENST00000336915.6
|
SKAP1
|
src kinase associated phosphoprotein 1 |
| chr19_+_37407351 | 0.15 |
ENST00000455427.2
ENST00000587857.1 |
ZNF568
|
zinc finger protein 568 |
| chr12_+_71833550 | 0.15 |
ENST00000266674.5
|
LGR5
|
leucine-rich repeat containing G protein-coupled receptor 5 |
| chr5_+_110407390 | 0.15 |
ENST00000344895.3
|
TSLP
|
thymic stromal lymphopoietin |
| chr3_+_177545563 | 0.15 |
ENST00000434309.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr2_+_232572361 | 0.15 |
ENST00000409321.1
|
PTMA
|
prothymosin, alpha |
| chr21_+_33671160 | 0.15 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr9_+_34458851 | 0.15 |
ENST00000545019.1
|
DNAI1
|
dynein, axonemal, intermediate chain 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX3-2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:2000974 | auditory receptor cell fate determination(GO:0042668) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.3 | 0.8 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.2 | 0.6 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 0.4 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.6 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.5 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.1 | 0.2 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.1 | 0.3 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.1 | 0.4 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.1 | 1.0 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.1 | 0.5 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.2 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.0 | 0.2 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.4 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.6 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.3 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.0 | 0.2 | GO:0070846 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.0 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 0.2 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 0.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.2 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.2 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.1 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.7 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.2 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.1 | GO:0032824 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.0 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.4 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.4 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.3 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.7 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.2 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.2 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.5 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.2 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.3 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 1.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.8 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.1 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.1 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.2 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.2 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0071351 | interleukin-18-mediated signaling pathway(GO:0035655) interleukin-33-mediated signaling pathway(GO:0038172) cellular response to interleukin-18(GO:0071351) |
| 0.0 | 0.3 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.4 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.1 | GO:0001828 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 1.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.2 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.2 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.2 | GO:1904936 | cerebral cortex tangential migration(GO:0021800) cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:2000690 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.0 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0002396 | MHC protein complex assembly(GO:0002396) MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.2 | GO:0090493 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.0 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.6 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.2 | GO:0060155 | platelet dense granule organization(GO:0060155) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.8 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.7 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.7 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.2 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.0 | 0.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.0 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 1.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.0 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.4 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 0.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.4 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.2 | GO:0005334 | norepinephrine:sodium symporter activity(GO:0005334) |
| 0.0 | 0.1 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 1.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 1.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.5 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 1.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.1 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.0 | 0.5 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.2 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.6 | GO:0016918 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.0 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.4 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.1 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.0 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.2 | GO:0055103 | ligase regulator activity(GO:0055103) |
| 0.0 | 0.1 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 2.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.0 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |