Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
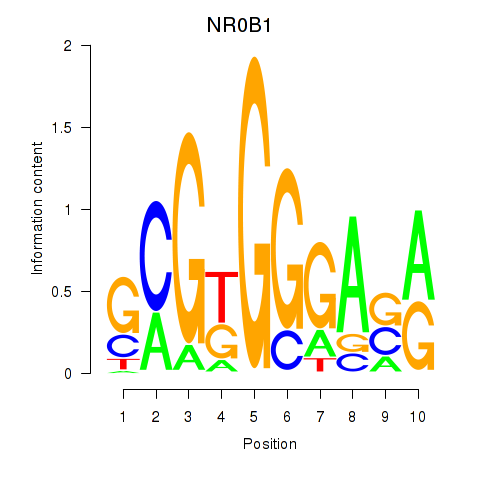
Results for NR0B1
Z-value: 0.60
Transcription factors associated with NR0B1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR0B1
|
ENSG00000169297.6 | nuclear receptor subfamily 0 group B member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR0B1 | hg19_v2_chrX_-_30327495_30327509 | -0.11 | 6.5e-01 | Click! |
Activity profile of NR0B1 motif
Sorted Z-values of NR0B1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_176915036 | 0.97 |
ENST00000427349.1
ENST00000352800.5 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr2_-_44223089 | 0.79 |
ENST00000447246.1
ENST00000409946.1 ENST00000409659.1 |
LRPPRC
|
leucine-rich pentatricopeptide repeat containing |
| chr13_-_52027134 | 0.78 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr11_-_73471655 | 0.73 |
ENST00000400470.2
|
RAB6A
|
RAB6A, member RAS oncogene family |
| chr17_+_29421987 | 0.58 |
ENST00000431387.4
|
NF1
|
neurofibromin 1 |
| chr5_-_142783694 | 0.58 |
ENST00000394466.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr1_+_203765437 | 0.56 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr3_-_113464906 | 0.54 |
ENST00000477813.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr11_-_61101247 | 0.52 |
ENST00000543627.1
|
DDB1
|
damage-specific DNA binding protein 1, 127kDa |
| chr17_+_29421900 | 0.50 |
ENST00000358273.4
ENST00000356175.3 |
NF1
|
neurofibromin 1 |
| chr12_+_110718428 | 0.50 |
ENST00000552636.1
|
ATP2A2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr2_+_170683979 | 0.47 |
ENST00000418381.1
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr11_-_12030746 | 0.47 |
ENST00000533813.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr9_+_128509624 | 0.45 |
ENST00000342287.5
ENST00000373487.4 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chrX_-_20284733 | 0.45 |
ENST00000438357.1
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr18_+_20494078 | 0.45 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr5_-_142784003 | 0.43 |
ENST00000416954.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr9_+_128509663 | 0.43 |
ENST00000373489.5
ENST00000373483.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr5_-_142784101 | 0.43 |
ENST00000503201.1
ENST00000502892.1 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr11_+_117015024 | 0.43 |
ENST00000530272.1
|
PAFAH1B2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 (30kDa) |
| chr21_+_40181520 | 0.41 |
ENST00000456966.1
|
ETS2
|
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
| chr11_-_12030681 | 0.40 |
ENST00000529338.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr10_+_70715884 | 0.40 |
ENST00000354185.4
|
DDX21
|
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr14_-_21905424 | 0.39 |
ENST00000553622.1
|
CHD8
|
chromodomain helicase DNA binding protein 8 |
| chr5_+_61708582 | 0.38 |
ENST00000325324.6
|
IPO11
|
importin 11 |
| chr2_+_201171577 | 0.38 |
ENST00000409397.2
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr18_+_29672573 | 0.38 |
ENST00000578107.1
ENST00000257190.5 ENST00000580499.1 |
RNF138
|
ring finger protein 138, E3 ubiquitin protein ligase |
| chr2_-_165698521 | 0.38 |
ENST00000409184.3
ENST00000392717.2 ENST00000456693.1 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr13_-_52026730 | 0.38 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr3_-_176914963 | 0.37 |
ENST00000450267.1
ENST00000431674.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr17_-_65241281 | 0.37 |
ENST00000358691.5
ENST00000580168.1 |
HELZ
|
helicase with zinc finger |
| chr17_+_36508826 | 0.37 |
ENST00000580660.1
|
SOCS7
|
suppressor of cytokine signaling 7 |
| chr5_+_138609782 | 0.36 |
ENST00000361059.2
ENST00000514694.1 ENST00000504203.1 ENST00000502929.1 ENST00000394800.2 ENST00000509644.1 ENST00000505016.1 |
MATR3
|
matrin 3 |
| chr11_-_2160180 | 0.36 |
ENST00000381406.4
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr3_+_111578583 | 0.36 |
ENST00000478922.1
ENST00000477695.1 |
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr8_-_142011291 | 0.36 |
ENST00000521059.1
|
PTK2
|
protein tyrosine kinase 2 |
| chrX_-_119695279 | 0.36 |
ENST00000336592.6
|
CUL4B
|
cullin 4B |
| chr8_-_142011244 | 0.36 |
ENST00000340930.3
ENST00000520828.1 ENST00000524257.1 ENST00000523679.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr3_-_45957088 | 0.36 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr3_+_111578131 | 0.35 |
ENST00000498699.1
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr16_-_18937726 | 0.35 |
ENST00000389467.3
ENST00000446231.2 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr8_-_142011316 | 0.35 |
ENST00000522684.1
ENST00000395218.2 ENST00000524357.1 ENST00000521332.1 ENST00000524040.1 ENST00000519881.1 ENST00000520045.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr2_+_209131059 | 0.34 |
ENST00000422495.1
ENST00000452564.1 |
PIKFYVE
|
phosphoinositide kinase, FYVE finger containing |
| chr2_+_201171064 | 0.34 |
ENST00000451764.2
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr8_-_67525473 | 0.34 |
ENST00000522677.3
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr2_+_201170999 | 0.34 |
ENST00000439395.1
ENST00000444012.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr12_-_6715808 | 0.34 |
ENST00000545584.1
|
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr1_-_46598371 | 0.34 |
ENST00000372006.1
ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr3_+_111578027 | 0.34 |
ENST00000431670.2
ENST00000412622.1 |
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr9_-_94877658 | 0.34 |
ENST00000262554.2
ENST00000337841.4 |
SPTLC1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr16_+_14280742 | 0.33 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr6_-_110500826 | 0.33 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr3_+_170075436 | 0.33 |
ENST00000476188.1
ENST00000259119.4 ENST00000426052.2 |
SKIL
|
SKI-like oncogene |
| chr1_-_93426998 | 0.33 |
ENST00000370310.4
|
FAM69A
|
family with sequence similarity 69, member A |
| chr17_-_4269768 | 0.33 |
ENST00000396981.2
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr9_-_134615326 | 0.33 |
ENST00000438647.1
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr10_-_69834973 | 0.32 |
ENST00000395187.2
|
HERC4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr5_+_138609441 | 0.32 |
ENST00000509990.1
ENST00000506147.1 ENST00000512107.1 |
MATR3
|
matrin 3 |
| chr17_+_45608614 | 0.32 |
ENST00000544660.1
|
NPEPPS
|
aminopeptidase puromycin sensitive |
| chr1_+_211433275 | 0.32 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr4_+_38665810 | 0.31 |
ENST00000261438.5
ENST00000514033.1 |
KLF3
|
Kruppel-like factor 3 (basic) |
| chr21_-_15755446 | 0.31 |
ENST00000544452.1
ENST00000285667.3 |
HSPA13
|
heat shock protein 70kDa family, member 13 |
| chr5_-_107717058 | 0.31 |
ENST00000359660.5
|
FBXL17
|
F-box and leucine-rich repeat protein 17 |
| chr20_+_2083540 | 0.31 |
ENST00000400064.3
|
STK35
|
serine/threonine kinase 35 |
| chr8_-_142011036 | 0.31 |
ENST00000520892.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr1_+_206138884 | 0.31 |
ENST00000341209.5
ENST00000607379.1 |
FAM72A
|
family with sequence similarity 72, member A |
| chr11_-_119993979 | 0.31 |
ENST00000524816.3
ENST00000525327.1 |
TRIM29
|
tripartite motif containing 29 |
| chr2_+_159313452 | 0.31 |
ENST00000389757.3
ENST00000389759.3 |
PKP4
|
plakophilin 4 |
| chr10_-_69835099 | 0.30 |
ENST00000373700.4
|
HERC4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr3_+_111578640 | 0.30 |
ENST00000393925.3
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr14_+_96968802 | 0.30 |
ENST00000556619.1
ENST00000392990.2 |
PAPOLA
|
poly(A) polymerase alpha |
| chr8_+_95653302 | 0.30 |
ENST00000423620.2
ENST00000433389.2 |
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr1_-_171711177 | 0.30 |
ENST00000415773.1
ENST00000367740.2 |
VAMP4
|
vesicle-associated membrane protein 4 |
| chr1_-_147245484 | 0.29 |
ENST00000271348.2
|
GJA5
|
gap junction protein, alpha 5, 40kDa |
| chr2_-_152684977 | 0.29 |
ENST00000428992.2
ENST00000295087.8 |
ARL5A
|
ADP-ribosylation factor-like 5A |
| chr2_-_44223138 | 0.29 |
ENST00000260665.7
|
LRPPRC
|
leucine-rich pentatricopeptide repeat containing |
| chr4_-_146101304 | 0.29 |
ENST00000447906.2
|
OTUD4
|
OTU domain containing 4 |
| chr12_-_39837192 | 0.29 |
ENST00000361961.3
ENST00000395670.3 |
KIF21A
|
kinesin family member 21A |
| chr11_+_117014983 | 0.29 |
ENST00000527958.1
ENST00000419197.2 ENST00000304808.6 ENST00000529887.2 |
PAFAH1B2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 (30kDa) |
| chr10_+_28821674 | 0.29 |
ENST00000526722.1
ENST00000375646.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr1_-_159915386 | 0.29 |
ENST00000361509.3
ENST00000368094.1 |
IGSF9
|
immunoglobulin superfamily, member 9 |
| chr6_-_111804393 | 0.28 |
ENST00000368802.3
ENST00000368805.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr10_-_123687431 | 0.28 |
ENST00000423243.1
|
ATE1
|
arginyltransferase 1 |
| chr15_-_66858298 | 0.27 |
ENST00000537670.1
|
LCTL
|
lactase-like |
| chr5_+_67511524 | 0.27 |
ENST00000521381.1
ENST00000521657.1 |
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr8_+_100025476 | 0.27 |
ENST00000355155.1
ENST00000357162.2 ENST00000358544.2 ENST00000395996.1 ENST00000441350.2 |
VPS13B
|
vacuolar protein sorting 13 homolog B (yeast) |
| chr12_-_57023995 | 0.27 |
ENST00000549884.1
ENST00000546695.1 |
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr12_+_27677085 | 0.26 |
ENST00000545334.1
ENST00000540114.1 ENST00000537927.1 ENST00000318304.8 ENST00000535047.1 ENST00000542629.1 ENST00000228425.6 |
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr8_-_67525524 | 0.26 |
ENST00000517885.1
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr15_-_91537723 | 0.26 |
ENST00000394249.3
ENST00000559811.1 ENST00000442656.2 ENST00000557905.1 ENST00000361919.3 |
PRC1
|
protein regulator of cytokinesis 1 |
| chrX_+_133507327 | 0.26 |
ENST00000332070.3
ENST00000394292.1 ENST00000370799.1 ENST00000416404.2 |
PHF6
|
PHD finger protein 6 |
| chr1_-_109584716 | 0.26 |
ENST00000531337.1
ENST00000529074.1 ENST00000369965.4 |
WDR47
|
WD repeat domain 47 |
| chr8_+_38614778 | 0.26 |
ENST00000521050.1
ENST00000522904.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr1_+_235492300 | 0.26 |
ENST00000476121.1
ENST00000497327.1 |
GGPS1
|
geranylgeranyl diphosphate synthase 1 |
| chrX_-_129402857 | 0.26 |
ENST00000447817.1
ENST00000370978.4 |
ZNF280C
|
zinc finger protein 280C |
| chr19_-_2050852 | 0.26 |
ENST00000541165.1
ENST00000591601.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr3_-_13921594 | 0.26 |
ENST00000285018.4
|
WNT7A
|
wingless-type MMTV integration site family, member 7A |
| chr2_+_170683942 | 0.26 |
ENST00000272793.5
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr9_+_131451480 | 0.25 |
ENST00000322030.8
|
SET
|
SET nuclear oncogene |
| chr19_-_51466681 | 0.25 |
ENST00000456750.2
|
KLK6
|
kallikrein-related peptidase 6 |
| chr3_-_64431058 | 0.25 |
ENST00000564377.1
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr6_-_94129244 | 0.25 |
ENST00000369303.4
ENST00000369297.1 |
EPHA7
|
EPH receptor A7 |
| chr11_+_57435441 | 0.25 |
ENST00000528177.1
|
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chrX_+_133507389 | 0.25 |
ENST00000370800.4
|
PHF6
|
PHD finger protein 6 |
| chr3_+_133292851 | 0.25 |
ENST00000503932.1
|
CDV3
|
CDV3 homolog (mouse) |
| chrX_-_131352152 | 0.24 |
ENST00000342983.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr1_-_54303949 | 0.24 |
ENST00000234725.8
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr4_-_151936416 | 0.24 |
ENST00000510413.1
ENST00000507224.1 |
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr12_+_41086215 | 0.24 |
ENST00000547702.1
ENST00000551424.1 |
CNTN1
|
contactin 1 |
| chr1_-_54303934 | 0.24 |
ENST00000537333.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr3_-_100120223 | 0.24 |
ENST00000284320.5
|
TOMM70A
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) |
| chr3_-_125094093 | 0.24 |
ENST00000484491.1
ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148
|
zinc finger protein 148 |
| chr6_-_111804905 | 0.24 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr10_+_5726764 | 0.24 |
ENST00000328090.5
ENST00000496681.1 |
FAM208B
|
family with sequence similarity 208, member B |
| chr18_-_29522989 | 0.24 |
ENST00000582539.1
ENST00000283351.4 ENST00000582513.1 |
TRAPPC8
|
trafficking protein particle complex 8 |
| chr2_-_183903133 | 0.24 |
ENST00000361354.4
|
NCKAP1
|
NCK-associated protein 1 |
| chr9_-_130966497 | 0.24 |
ENST00000393608.1
ENST00000372948.3 |
CIZ1
|
CDKN1A interacting zinc finger protein 1 |
| chr13_+_76210448 | 0.24 |
ENST00000377499.5
|
LMO7
|
LIM domain 7 |
| chr8_-_101964231 | 0.24 |
ENST00000521309.1
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr2_+_14772810 | 0.23 |
ENST00000295092.2
ENST00000331243.4 |
FAM84A
|
family with sequence similarity 84, member A |
| chr15_-_83953466 | 0.23 |
ENST00000345382.2
|
BNC1
|
basonuclin 1 |
| chr19_-_4722780 | 0.23 |
ENST00000600621.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr8_-_42396721 | 0.23 |
ENST00000518384.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr5_+_43121698 | 0.23 |
ENST00000505606.2
ENST00000509634.1 ENST00000509341.1 |
ZNF131
|
zinc finger protein 131 |
| chr17_-_27181990 | 0.23 |
ENST00000583953.1
|
FAM222B
|
family with sequence similarity 222, member B |
| chr14_+_73525229 | 0.23 |
ENST00000527432.1
ENST00000531500.1 ENST00000525321.1 ENST00000526754.1 |
RBM25
|
RNA binding motif protein 25 |
| chr3_+_73045936 | 0.23 |
ENST00000356692.5
ENST00000488810.1 ENST00000394284.3 ENST00000295862.9 ENST00000495566.1 |
PPP4R2
|
protein phosphatase 4, regulatory subunit 2 |
| chr2_-_165698322 | 0.23 |
ENST00000444537.1
ENST00000414843.1 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr22_+_29876197 | 0.22 |
ENST00000310624.6
|
NEFH
|
neurofilament, heavy polypeptide |
| chr10_-_112064665 | 0.22 |
ENST00000369603.5
|
SMNDC1
|
survival motor neuron domain containing 1 |
| chr1_+_95699704 | 0.22 |
ENST00000370202.4
|
RWDD3
|
RWD domain containing 3 |
| chr21_+_17102311 | 0.22 |
ENST00000285679.6
ENST00000351097.5 ENST00000285681.2 ENST00000400183.2 |
USP25
|
ubiquitin specific peptidase 25 |
| chr12_-_120554534 | 0.22 |
ENST00000538903.1
ENST00000534951.1 |
RAB35
|
RAB35, member RAS oncogene family |
| chr12_-_6484715 | 0.22 |
ENST00000228916.2
|
SCNN1A
|
sodium channel, non-voltage-gated 1 alpha subunit |
| chr20_+_57466629 | 0.22 |
ENST00000371081.1
ENST00000338783.6 |
GNAS
|
GNAS complex locus |
| chr5_-_77590480 | 0.22 |
ENST00000519295.1
ENST00000255194.6 |
AP3B1
|
adaptor-related protein complex 3, beta 1 subunit |
| chr1_-_171711387 | 0.22 |
ENST00000236192.7
|
VAMP4
|
vesicle-associated membrane protein 4 |
| chr21_-_34100244 | 0.22 |
ENST00000382491.3
ENST00000357345.3 ENST00000429236.1 |
SYNJ1
|
synaptojanin 1 |
| chr8_-_101734308 | 0.21 |
ENST00000519004.1
ENST00000519363.1 ENST00000520142.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr1_-_156390128 | 0.21 |
ENST00000368242.3
|
C1orf61
|
chromosome 1 open reading frame 61 |
| chrX_+_133507283 | 0.21 |
ENST00000370803.3
|
PHF6
|
PHD finger protein 6 |
| chr13_+_25670268 | 0.21 |
ENST00000281589.3
|
PABPC3
|
poly(A) binding protein, cytoplasmic 3 |
| chr20_+_35807714 | 0.21 |
ENST00000373632.4
|
RPN2
|
ribophorin II |
| chr2_+_217277137 | 0.21 |
ENST00000430374.1
ENST00000357276.4 ENST00000444508.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr16_+_14280564 | 0.21 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr14_+_57046530 | 0.21 |
ENST00000536419.1
ENST00000538838.1 |
TMEM260
|
transmembrane protein 260 |
| chr1_-_197169672 | 0.21 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chrX_-_129244336 | 0.20 |
ENST00000434609.1
|
ELF4
|
E74-like factor 4 (ets domain transcription factor) |
| chr20_-_46415341 | 0.20 |
ENST00000484875.1
ENST00000361612.4 |
SULF2
|
sulfatase 2 |
| chr14_+_57857262 | 0.20 |
ENST00000555166.1
ENST00000556492.1 ENST00000554703.1 |
NAA30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr11_-_92930556 | 0.20 |
ENST00000529184.1
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr5_+_112312454 | 0.20 |
ENST00000543319.1
|
DCP2
|
decapping mRNA 2 |
| chr5_+_112312399 | 0.20 |
ENST00000515408.1
ENST00000513585.1 |
DCP2
|
decapping mRNA 2 |
| chr4_+_159593418 | 0.20 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr21_-_34100341 | 0.20 |
ENST00000382499.2
ENST00000433931.2 |
SYNJ1
|
synaptojanin 1 |
| chr6_-_160210604 | 0.20 |
ENST00000420894.2
ENST00000539756.1 ENST00000544255.1 |
TCP1
|
t-complex 1 |
| chr10_-_21463116 | 0.19 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chr7_-_151329416 | 0.19 |
ENST00000418337.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chrX_+_9754461 | 0.19 |
ENST00000380913.3
|
SHROOM2
|
shroom family member 2 |
| chr1_-_46598284 | 0.19 |
ENST00000423209.1
ENST00000262741.5 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr8_-_27469383 | 0.19 |
ENST00000519742.1
|
CLU
|
clusterin |
| chr9_+_79792269 | 0.19 |
ENST00000376634.4
ENST00000376636.3 ENST00000360280.3 |
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae) |
| chr1_-_147245445 | 0.19 |
ENST00000430508.1
|
GJA5
|
gap junction protein, alpha 5, 40kDa |
| chr7_+_99156314 | 0.19 |
ENST00000425063.1
ENST00000493277.1 |
ZNF655
|
zinc finger protein 655 |
| chr11_+_117103441 | 0.19 |
ENST00000531287.1
ENST00000531452.1 |
RNF214
|
ring finger protein 214 |
| chr5_-_137368726 | 0.19 |
ENST00000420893.2
ENST00000425075.2 |
FAM13B
|
family with sequence similarity 13, member B |
| chr20_-_46415297 | 0.19 |
ENST00000467815.1
ENST00000359930.4 |
SULF2
|
sulfatase 2 |
| chr1_+_180601139 | 0.19 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr17_-_79817091 | 0.18 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr3_-_33260707 | 0.18 |
ENST00000309558.3
|
SUSD5
|
sushi domain containing 5 |
| chr16_-_69166031 | 0.18 |
ENST00000398235.2
ENST00000520529.1 |
CHTF8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae) |
| chr12_+_69864129 | 0.18 |
ENST00000547219.1
ENST00000299293.2 ENST00000549921.1 ENST00000550316.1 ENST00000548154.1 ENST00000547414.1 ENST00000550389.1 ENST00000550937.1 ENST00000549092.1 ENST00000550169.1 |
FRS2
|
fibroblast growth factor receptor substrate 2 |
| chr17_-_48785216 | 0.18 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr11_+_35684288 | 0.18 |
ENST00000299413.5
|
TRIM44
|
tripartite motif containing 44 |
| chr8_+_125486939 | 0.17 |
ENST00000303545.3
|
RNF139
|
ring finger protein 139 |
| chr11_-_12030629 | 0.17 |
ENST00000396505.2
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr22_+_19702069 | 0.17 |
ENST00000412544.1
|
SEPT5
|
septin 5 |
| chr10_+_31608054 | 0.17 |
ENST00000320985.10
ENST00000361642.5 ENST00000560721.2 ENST00000558440.1 ENST00000424869.1 ENST00000542815.3 |
ZEB1
|
zinc finger E-box binding homeobox 1 |
| chr6_+_7541808 | 0.17 |
ENST00000379802.3
|
DSP
|
desmoplakin |
| chr15_-_51914810 | 0.17 |
ENST00000543779.2
ENST00000449909.3 |
DMXL2
|
Dmx-like 2 |
| chr1_-_54304212 | 0.17 |
ENST00000540001.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr14_+_73525144 | 0.17 |
ENST00000261973.7
ENST00000540173.1 |
RBM25
|
RNA binding motif protein 25 |
| chr1_+_52608046 | 0.17 |
ENST00000357206.2
ENST00000287727.3 |
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr12_-_96794143 | 0.16 |
ENST00000543119.2
|
CDK17
|
cyclin-dependent kinase 17 |
| chr6_+_17393888 | 0.16 |
ENST00000493172.1
ENST00000465994.1 |
CAP2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr6_+_7541845 | 0.15 |
ENST00000418664.2
|
DSP
|
desmoplakin |
| chr2_-_201828356 | 0.15 |
ENST00000234296.2
|
ORC2
|
origin recognition complex, subunit 2 |
| chr1_-_236030216 | 0.15 |
ENST00000389794.3
ENST00000389793.2 |
LYST
|
lysosomal trafficking regulator |
| chr1_-_94312706 | 0.15 |
ENST00000370244.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr17_-_28619059 | 0.15 |
ENST00000580709.1
|
BLMH
|
bleomycin hydrolase |
| chr19_-_4722705 | 0.15 |
ENST00000598360.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr14_+_96968707 | 0.15 |
ENST00000216277.8
ENST00000557320.1 ENST00000557471.1 |
PAPOLA
|
poly(A) polymerase alpha |
| chr1_-_42800614 | 0.15 |
ENST00000372572.1
|
FOXJ3
|
forkhead box J3 |
| chr1_-_183604794 | 0.15 |
ENST00000367534.1
ENST00000359856.6 ENST00000294742.6 |
ARPC5
|
actin related protein 2/3 complex, subunit 5, 16kDa |
| chr12_+_109535373 | 0.15 |
ENST00000242576.2
|
UNG
|
uracil-DNA glycosylase |
| chr16_+_56764017 | 0.15 |
ENST00000308159.5
ENST00000563858.1 ENST00000566315.1 ENST00000569842.1 ENST00000569863.1 |
NUP93
|
nucleoporin 93kDa |
| chr6_+_116692102 | 0.15 |
ENST00000359564.2
|
DSE
|
dermatan sulfate epimerase |
| chr6_+_7107830 | 0.14 |
ENST00000379933.3
|
RREB1
|
ras responsive element binding protein 1 |
| chr1_+_211432700 | 0.14 |
ENST00000452621.2
|
RCOR3
|
REST corepressor 3 |
| chr4_+_88928777 | 0.14 |
ENST00000237596.2
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant) |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR0B1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.4 | 1.1 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.3 | 0.9 | GO:0007388 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.2 | 0.7 | GO:0030474 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.2 | 0.5 | GO:0086054 | atrial ventricular junction remodeling(GO:0003294) atrial cardiac muscle cell to AV node cell communication by electrical coupling(GO:0086044) bundle of His cell to Purkinje myocyte communication by electrical coupling(GO:0086054) Purkinje myocyte to ventricular cardiac muscle cell communication by electrical coupling(GO:0086055) regulation of Purkinje myocyte action potential(GO:0098906) vasomotion(GO:1990029) |
| 0.2 | 0.5 | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum(GO:1903515) |
| 0.1 | 0.7 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.7 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 1.0 | GO:1902612 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.1 | 0.4 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.1 | 0.4 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.3 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.1 | 0.4 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.3 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.5 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 0.4 | GO:1904978 | regulation of endosome organization(GO:1904978) |
| 0.1 | 1.4 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.4 | GO:0051808 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.1 | 0.4 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.1 | 1.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 1.3 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.3 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 1.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.2 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 0.2 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.3 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.4 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.2 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.1 | 0.4 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.2 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.2 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.1 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 | 0.4 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.5 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 1.1 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.4 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.2 | GO:0051621 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.2 | GO:0001738 | morphogenesis of a polarized epithelium(GO:0001738) |
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.3 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.2 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.2 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 0.5 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.4 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.3 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.4 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.3 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:0090096 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 | 0.1 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.0 | 0.8 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.6 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.2 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.2 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.1 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 | 0.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.0 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 0.1 | GO:1904799 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.2 | GO:0046598 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.0 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0060623 | pericentric heterochromatin assembly(GO:0031508) regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.0 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.0 | 0.2 | GO:0090520 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.4 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.5 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.0 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.2 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.1 | 0.7 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 1.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.4 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 1.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.2 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.5 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 1.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 2.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.4 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.0 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0086079 | gap junction channel activity involved in atrial cardiac muscle cell-AV node cell electrical coupling(GO:0086076) gap junction channel activity involved in bundle of His cell-Purkinje myocyte electrical coupling(GO:0086078) gap junction channel activity involved in Purkinje myocyte-ventricular cardiac muscle cell electrical coupling(GO:0086079) |
| 0.2 | 1.4 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.2 | 0.5 | GO:0031775 | lutropin-choriogonadotropic hormone receptor binding(GO:0031775) calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential(GO:0086039) |
| 0.1 | 0.7 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 0.8 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.1 | 0.4 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 0.1 | 0.4 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.2 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 1.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.2 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.4 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.0 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.1 | GO:0048763 | HLH domain binding(GO:0043398) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.3 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.0 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 1.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.2 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 1.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.3 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.9 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.7 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 2.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |