Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
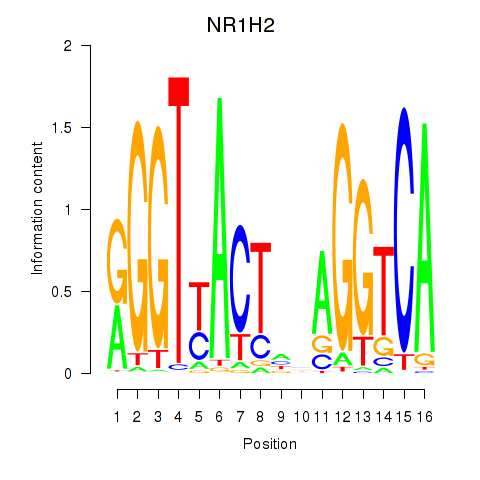
Results for NR1H2
Z-value: 1.38
Transcription factors associated with NR1H2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1H2
|
ENSG00000131408.9 | nuclear receptor subfamily 1 group H member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1H2 | hg19_v2_chr19_+_50879705_50879775 | 0.96 | 2.2e-11 | Click! |
Activity profile of NR1H2 motif
Sorted Z-values of NR1H2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_124749609 | 5.70 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr17_-_19648916 | 4.50 |
ENST00000444455.1
ENST00000439102.2 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr20_+_43343886 | 4.08 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr19_-_38747172 | 3.86 |
ENST00000347262.4
ENST00000591585.1 ENST00000301242.4 |
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr14_-_102976135 | 3.78 |
ENST00000560748.1
|
ANKRD9
|
ankyrin repeat domain 9 |
| chr20_+_43343517 | 3.40 |
ENST00000372865.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr2_-_216946500 | 3.38 |
ENST00000265322.7
|
PECR
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr17_+_77019030 | 3.35 |
ENST00000580454.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr20_+_34679725 | 3.22 |
ENST00000432589.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr17_-_76183111 | 3.09 |
ENST00000405273.1
ENST00000590862.1 ENST00000590430.1 ENST00000586613.1 |
TK1
|
thymidine kinase 1, soluble |
| chr19_-_38806540 | 3.09 |
ENST00000592694.1
|
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr11_+_844406 | 3.08 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr8_-_61193947 | 2.80 |
ENST00000317995.4
|
CA8
|
carbonic anhydrase VIII |
| chr8_-_48651648 | 2.78 |
ENST00000408965.3
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr17_-_17480779 | 2.76 |
ENST00000395782.1
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr17_+_77018896 | 2.75 |
ENST00000578229.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr17_-_26695013 | 2.64 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr16_-_2205352 | 2.57 |
ENST00000563192.1
|
RP11-304L19.5
|
RP11-304L19.5 |
| chr2_+_223725723 | 2.55 |
ENST00000535678.1
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr6_+_30687978 | 2.40 |
ENST00000327892.8
ENST00000435534.1 |
TUBB
|
tubulin, beta class I |
| chr17_-_26694979 | 2.40 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr19_+_41092680 | 2.39 |
ENST00000594298.1
ENST00000597396.1 |
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr19_+_15751689 | 2.38 |
ENST00000586182.2
ENST00000591058.1 ENST00000221307.8 |
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3 |
| chr2_-_74699770 | 2.37 |
ENST00000409710.1
|
MRPL53
|
mitochondrial ribosomal protein L53 |
| chr19_+_2977444 | 2.36 |
ENST00000246112.4
ENST00000453329.1 ENST00000482627.1 ENST00000452088.1 |
TLE6
|
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
| chr15_-_63674218 | 2.35 |
ENST00000178638.3
|
CA12
|
carbonic anhydrase XII |
| chr20_+_5987890 | 2.35 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr19_-_38806560 | 2.30 |
ENST00000591755.1
ENST00000337679.8 ENST00000339413.6 |
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr19_-_38806390 | 2.26 |
ENST00000589247.1
ENST00000329420.8 ENST00000591784.1 |
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr12_+_22778009 | 2.19 |
ENST00000266517.4
ENST00000335148.3 |
ETNK1
|
ethanolamine kinase 1 |
| chr19_-_46285646 | 2.16 |
ENST00000458663.2
|
DMPK
|
dystrophia myotonica-protein kinase |
| chr20_-_32262165 | 2.11 |
ENST00000606690.1
ENST00000246190.6 ENST00000439478.1 ENST00000375238.4 |
NECAB3
|
N-terminal EF-hand calcium binding protein 3 |
| chr16_+_577697 | 2.05 |
ENST00000562370.1
ENST00000568988.1 ENST00000219611.2 |
CAPN15
|
calpain 15 |
| chr2_+_228029281 | 2.03 |
ENST00000396578.3
|
COL4A3
|
collagen, type IV, alpha 3 (Goodpasture antigen) |
| chr19_+_15752088 | 1.97 |
ENST00000585846.1
|
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3 |
| chr5_-_58652788 | 1.95 |
ENST00000405755.2
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr19_+_34287174 | 1.93 |
ENST00000587559.1
ENST00000588637.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr2_+_216946589 | 1.90 |
ENST00000433112.1
ENST00000454545.1 ENST00000437356.2 ENST00000295658.4 ENST00000455479.1 ENST00000406027.2 |
TMEM169
|
transmembrane protein 169 |
| chrX_+_1455478 | 1.87 |
ENST00000331035.4
|
IL3RA
|
interleukin 3 receptor, alpha (low affinity) |
| chr20_+_34680595 | 1.86 |
ENST00000406771.2
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr1_+_169075554 | 1.83 |
ENST00000367815.4
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr3_-_49893958 | 1.78 |
ENST00000482243.1
ENST00000331456.2 ENST00000469027.1 |
TRAIP
|
TRAF interacting protein |
| chr19_-_17622684 | 1.78 |
ENST00000596643.1
|
CTD-3131K8.2
|
CTD-3131K8.2 |
| chr11_+_62652649 | 1.75 |
ENST00000539507.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr15_-_102285007 | 1.75 |
ENST00000560292.2
|
RP11-89K11.1
|
Uncharacterized protein |
| chr11_+_65190245 | 1.70 |
ENST00000499732.1
ENST00000501122.2 ENST00000601801.1 |
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr16_-_30023615 | 1.65 |
ENST00000564979.1
ENST00000563378.1 |
DOC2A
|
double C2-like domains, alpha |
| chr15_+_89010923 | 1.60 |
ENST00000353598.6
|
MRPS11
|
mitochondrial ribosomal protein S11 |
| chr14_-_102976091 | 1.60 |
ENST00000286918.4
|
ANKRD9
|
ankyrin repeat domain 9 |
| chr1_+_10459111 | 1.60 |
ENST00000541529.1
ENST00000270776.8 ENST00000483936.1 ENST00000538557.1 |
PGD
|
phosphogluconate dehydrogenase |
| chr12_+_22778116 | 1.60 |
ENST00000538218.1
|
ETNK1
|
ethanolamine kinase 1 |
| chr20_+_34680620 | 1.55 |
ENST00000430276.1
ENST00000373950.2 ENST00000452261.1 |
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr8_+_142402089 | 1.51 |
ENST00000521578.1
ENST00000520105.1 ENST00000523147.1 |
PTP4A3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr17_+_42977122 | 1.45 |
ENST00000412523.2
ENST00000331733.4 ENST00000417826.2 |
FAM187A
CCDC103
|
family with sequence similarity 187, member A coiled-coil domain containing 103 |
| chr1_-_153950098 | 1.40 |
ENST00000356648.1
|
JTB
|
jumping translocation breakpoint |
| chr17_+_4675175 | 1.37 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr19_+_33071974 | 1.32 |
ENST00000590247.2
ENST00000419343.3 ENST00000592786.1 ENST00000379316.3 |
PDCD5
|
programmed cell death 5 |
| chr19_+_15783879 | 1.29 |
ENST00000551607.1
|
CYP4F12
|
cytochrome P450, family 4, subfamily F, polypeptide 12 |
| chrX_+_52240504 | 1.28 |
ENST00000399805.2
|
XAGE1B
|
X antigen family, member 1B |
| chr4_-_140216948 | 1.25 |
ENST00000265500.4
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr9_+_67977438 | 1.25 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr12_+_57998400 | 1.24 |
ENST00000548804.1
ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr12_+_122064398 | 1.21 |
ENST00000330079.7
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr10_-_72648541 | 1.20 |
ENST00000299299.3
|
PCBD1
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
| chr15_-_89010607 | 1.19 |
ENST00000312475.4
|
MRPL46
|
mitochondrial ribosomal protein L46 |
| chr19_-_49121054 | 1.17 |
ENST00000546623.1
ENST00000084795.5 |
RPL18
|
ribosomal protein L18 |
| chrX_+_102631248 | 1.15 |
ENST00000361298.4
ENST00000372645.3 ENST00000372635.1 |
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr16_-_46649905 | 1.14 |
ENST00000569702.1
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chr14_-_93673353 | 1.12 |
ENST00000556566.1
ENST00000306954.4 |
C14orf142
|
chromosome 14 open reading frame 142 |
| chr7_+_100136811 | 1.11 |
ENST00000300176.4
ENST00000262935.4 |
AGFG2
|
ArfGAP with FG repeats 2 |
| chr16_-_4465886 | 1.11 |
ENST00000539968.1
|
CORO7
|
coronin 7 |
| chr17_+_70026795 | 1.10 |
ENST00000472655.2
ENST00000538810.1 |
RP11-84E24.3
|
long intergenic non-protein coding RNA 1152 |
| chr7_-_73153161 | 1.10 |
ENST00000395147.4
|
ABHD11
|
abhydrolase domain containing 11 |
| chr11_-_61596753 | 1.07 |
ENST00000448607.1
ENST00000421879.1 |
FADS1
|
fatty acid desaturase 1 |
| chr2_+_73441350 | 1.07 |
ENST00000389501.4
|
SMYD5
|
SMYD family member 5 |
| chr17_-_8151353 | 1.06 |
ENST00000315684.8
|
CTC1
|
CTS telomere maintenance complex component 1 |
| chrX_-_107334790 | 1.06 |
ENST00000217958.3
|
PSMD10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr17_-_8198636 | 1.05 |
ENST00000577745.1
ENST00000579192.1 ENST00000396278.1 |
SLC25A35
|
solute carrier family 25, member 35 |
| chr7_-_73153122 | 1.05 |
ENST00000458339.1
|
ABHD11
|
abhydrolase domain containing 11 |
| chr19_+_17865011 | 1.05 |
ENST00000596462.1
ENST00000596865.1 ENST00000598960.1 ENST00000539407.1 |
FCHO1
|
FCH domain only 1 |
| chr3_-_127309550 | 1.04 |
ENST00000296210.7
ENST00000355552.3 |
TPRA1
|
transmembrane protein, adipocyte asscociated 1 |
| chr14_-_59950724 | 1.00 |
ENST00000481608.1
|
L3HYPDH
|
L-3-hydroxyproline dehydratase (trans-) |
| chr1_+_45274154 | 0.97 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr1_-_153919128 | 0.97 |
ENST00000361217.4
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr19_+_1077393 | 0.95 |
ENST00000590577.1
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chr12_-_91573132 | 0.95 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr19_+_17622415 | 0.95 |
ENST00000252603.2
ENST00000600923.1 |
PGLS
|
6-phosphogluconolactonase |
| chr19_+_39881951 | 0.95 |
ENST00000315588.5
ENST00000594368.1 ENST00000599213.2 ENST00000596297.1 |
MED29
|
mediator complex subunit 29 |
| chr19_+_50094866 | 0.90 |
ENST00000418929.2
|
PRR12
|
proline rich 12 |
| chr22_-_24096630 | 0.90 |
ENST00000248948.3
|
VPREB3
|
pre-B lymphocyte 3 |
| chr2_+_44502597 | 0.89 |
ENST00000260649.6
ENST00000409387.1 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr19_-_36391434 | 0.89 |
ENST00000396901.1
ENST00000585925.1 |
NFKBID
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta |
| chr10_-_70287172 | 0.88 |
ENST00000539557.1
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr4_-_84205905 | 0.87 |
ENST00000311461.7
ENST00000311469.4 ENST00000439031.2 |
COQ2
|
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
| chr1_-_153949751 | 0.87 |
ENST00000428469.1
|
JTB
|
jumping translocation breakpoint |
| chr15_+_75498739 | 0.86 |
ENST00000565074.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chrX_-_107334750 | 0.84 |
ENST00000340200.5
ENST00000372296.1 ENST00000372295.1 ENST00000361815.5 |
PSMD10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr17_+_48351785 | 0.83 |
ENST00000507382.1
|
TMEM92
|
transmembrane protein 92 |
| chr17_-_41977964 | 0.83 |
ENST00000377184.3
|
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr8_-_145652336 | 0.82 |
ENST00000529182.1
ENST00000526054.1 |
VPS28
|
vacuolar protein sorting 28 homolog (S. cerevisiae) |
| chr5_-_133968529 | 0.82 |
ENST00000402673.2
|
SAR1B
|
SAR1 homolog B (S. cerevisiae) |
| chr11_+_86748863 | 0.81 |
ENST00000340353.7
|
TMEM135
|
transmembrane protein 135 |
| chr4_+_183164574 | 0.78 |
ENST00000511685.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr19_+_56146352 | 0.78 |
ENST00000592881.1
|
ZNF580
|
zinc finger protein 580 |
| chr16_-_1661988 | 0.78 |
ENST00000426508.2
|
IFT140
|
intraflagellar transport 140 homolog (Chlamydomonas) |
| chr16_+_72042487 | 0.76 |
ENST00000572887.1
ENST00000219240.4 ENST00000574309.1 ENST00000576145.1 |
DHODH
|
dihydroorotate dehydrogenase (quinone) |
| chr9_-_98189055 | 0.76 |
ENST00000433644.2
|
RP11-435O5.2
|
RP11-435O5.2 |
| chr17_-_27054952 | 0.75 |
ENST00000580518.1
|
TLCD1
|
TLC domain containing 1 |
| chr1_-_40098672 | 0.75 |
ENST00000535435.1
|
HEYL
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr12_+_100594557 | 0.74 |
ENST00000546902.1
ENST00000552376.1 ENST00000551617.1 |
ACTR6
|
ARP6 actin-related protein 6 homolog (yeast) |
| chr19_+_3224700 | 0.73 |
ENST00000292672.2
ENST00000541430.2 |
CELF5
|
CUGBP, Elav-like family member 5 |
| chr22_-_38480100 | 0.71 |
ENST00000427592.1
|
SLC16A8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr3_-_127309879 | 0.71 |
ENST00000489960.1
ENST00000490290.1 |
TPRA1
|
transmembrane protein, adipocyte asscociated 1 |
| chr17_+_34087888 | 0.70 |
ENST00000586491.1
ENST00000588628.1 ENST00000285023.4 |
C17orf50
|
chromosome 17 open reading frame 50 |
| chr11_+_827248 | 0.69 |
ENST00000527089.1
ENST00000530183.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr2_-_32390801 | 0.69 |
ENST00000608489.1
|
RP11-563N4.1
|
RP11-563N4.1 |
| chr1_-_201476220 | 0.68 |
ENST00000526723.1
ENST00000524951.1 |
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr14_-_70826438 | 0.65 |
ENST00000389912.6
|
COX16
|
COX16 cytochrome c oxidase assembly homolog (S. cerevisiae) |
| chr22_-_38577782 | 0.65 |
ENST00000430886.1
ENST00000332509.3 ENST00000447598.2 ENST00000435484.1 ENST00000402064.1 ENST00000436218.1 |
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr14_+_77787227 | 0.65 |
ENST00000216465.5
ENST00000361389.4 ENST00000554279.1 ENST00000557639.1 ENST00000349555.3 ENST00000556627.1 ENST00000557053.1 |
GSTZ1
|
glutathione S-transferase zeta 1 |
| chr17_-_3595042 | 0.64 |
ENST00000552723.1
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr22_-_24096562 | 0.64 |
ENST00000398465.3
|
VPREB3
|
pre-B lymphocyte 3 |
| chr3_+_52007693 | 0.63 |
ENST00000494478.1
|
ABHD14A
|
abhydrolase domain containing 14A |
| chr22_-_23974506 | 0.62 |
ENST00000317749.5
|
C22orf43
|
chromosome 22 open reading frame 43 |
| chr2_+_145425573 | 0.61 |
ENST00000600064.1
ENST00000597670.1 ENST00000414256.1 ENST00000599187.1 ENST00000451774.1 ENST00000599072.1 ENST00000596589.1 ENST00000597893.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr22_+_19118321 | 0.61 |
ENST00000399635.2
|
TSSK2
|
testis-specific serine kinase 2 |
| chr12_-_91573249 | 0.61 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr19_+_33622996 | 0.57 |
ENST00000592765.1
ENST00000361680.2 ENST00000355868.3 |
WDR88
|
WD repeat domain 88 |
| chr14_-_80782219 | 0.56 |
ENST00000553594.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr11_+_119039414 | 0.56 |
ENST00000409991.1
ENST00000292199.2 ENST00000409265.4 ENST00000409109.1 |
NLRX1
|
NLR family member X1 |
| chr1_+_6615241 | 0.56 |
ENST00000333172.6
ENST00000328191.4 ENST00000351136.3 |
TAS1R1
|
taste receptor, type 1, member 1 |
| chr11_-_82611448 | 0.56 |
ENST00000393399.2
ENST00000313010.3 |
PRCP
|
prolylcarboxypeptidase (angiotensinase C) |
| chr9_-_36400213 | 0.54 |
ENST00000259605.6
ENST00000353739.4 |
RNF38
|
ring finger protein 38 |
| chrX_+_49020121 | 0.54 |
ENST00000415364.1
ENST00000376338.3 ENST00000425285.1 |
MAGIX
|
MAGI family member, X-linked |
| chr11_+_10326612 | 0.54 |
ENST00000534464.1
ENST00000530439.1 ENST00000524948.1 ENST00000528655.1 ENST00000526492.1 ENST00000525063.1 |
ADM
|
adrenomedullin |
| chr4_-_83483395 | 0.51 |
ENST00000515780.2
|
TMEM150C
|
transmembrane protein 150C |
| chr22_-_33968239 | 0.49 |
ENST00000452586.2
ENST00000421768.1 |
LARGE
|
like-glycosyltransferase |
| chr22_-_38577731 | 0.49 |
ENST00000335539.3
|
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr11_-_68519026 | 0.49 |
ENST00000255087.5
|
MTL5
|
metallothionein-like 5, testis-specific (tesmin) |
| chr11_-_798239 | 0.49 |
ENST00000531437.1
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr17_-_15370915 | 0.49 |
ENST00000312177.6
|
CDRT4
|
CMT1A duplicated region transcript 4 |
| chr1_+_100818009 | 0.48 |
ENST00000370125.2
ENST00000361544.6 ENST00000370124.3 |
CDC14A
|
cell division cycle 14A |
| chr2_+_32390925 | 0.46 |
ENST00000440718.1
ENST00000379343.2 ENST00000282587.5 ENST00000435660.1 ENST00000538303.1 ENST00000357055.3 ENST00000406369.1 |
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr8_-_93115445 | 0.46 |
ENST00000523629.1
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr12_-_6677422 | 0.44 |
ENST00000382421.3
ENST00000545200.1 ENST00000399466.2 ENST00000536124.1 ENST00000540228.1 ENST00000542867.1 ENST00000545492.1 ENST00000322166.5 ENST00000545915.1 |
NOP2
|
NOP2 nucleolar protein |
| chr15_+_91473403 | 0.44 |
ENST00000394275.2
|
UNC45A
|
unc-45 homolog A (C. elegans) |
| chr11_+_61159832 | 0.42 |
ENST00000334888.5
ENST00000398979.3 |
TMEM216
|
transmembrane protein 216 |
| chr20_+_35918035 | 0.42 |
ENST00000373606.3
ENST00000397156.3 ENST00000397150.1 ENST00000397152.3 |
MANBAL
|
mannosidase, beta A, lysosomal-like |
| chr16_+_4674787 | 0.41 |
ENST00000262370.7
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr10_+_101491968 | 0.41 |
ENST00000370476.5
ENST00000370472.4 |
CUTC
|
cutC copper transporter |
| chr15_+_74610894 | 0.41 |
ENST00000558821.1
ENST00000268082.4 |
CCDC33
|
coiled-coil domain containing 33 |
| chr5_+_446253 | 0.39 |
ENST00000315013.5
|
EXOC3
|
exocyst complex component 3 |
| chr3_-_49466686 | 0.39 |
ENST00000273598.3
ENST00000436744.2 |
NICN1
|
nicolin 1 |
| chr16_+_4674814 | 0.39 |
ENST00000415496.1
ENST00000587747.1 ENST00000399577.5 ENST00000588994.1 ENST00000586183.1 |
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr7_+_2687173 | 0.38 |
ENST00000403167.1
|
TTYH3
|
tweety family member 3 |
| chr17_+_60457914 | 0.38 |
ENST00000305286.3
ENST00000520404.1 ENST00000518576.1 |
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr16_+_29467780 | 0.38 |
ENST00000395400.3
|
SULT1A4
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4 |
| chr16_-_14788526 | 0.38 |
ENST00000438167.3
|
PLA2G10
|
phospholipase A2, group X |
| chr2_+_145425534 | 0.37 |
ENST00000432608.1
ENST00000597655.1 ENST00000598659.1 ENST00000600679.1 ENST00000601277.1 ENST00000451027.1 ENST00000445791.1 ENST00000596540.1 ENST00000596230.1 ENST00000594471.1 ENST00000598248.1 ENST00000597469.1 ENST00000431734.1 ENST00000595686.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr11_-_68518910 | 0.37 |
ENST00000544963.1
ENST00000443940.2 |
MTL5
|
metallothionein-like 5, testis-specific (tesmin) |
| chr12_-_56122761 | 0.37 |
ENST00000552164.1
ENST00000420846.3 ENST00000257857.4 |
CD63
|
CD63 molecule |
| chr19_-_50420918 | 0.36 |
ENST00000595761.1
|
NUP62
|
nucleoporin 62kDa |
| chr3_-_114173530 | 0.36 |
ENST00000470311.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr11_-_62420757 | 0.36 |
ENST00000330574.2
|
INTS5
|
integrator complex subunit 5 |
| chr19_-_46296011 | 0.36 |
ENST00000377735.3
ENST00000270223.6 |
DMWD
|
dystrophia myotonica, WD repeat containing |
| chr14_-_22005062 | 0.36 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr16_+_29973351 | 0.35 |
ENST00000602948.1
ENST00000279396.6 ENST00000575829.2 ENST00000561899.2 |
TMEM219
|
transmembrane protein 219 |
| chr2_+_207630081 | 0.35 |
ENST00000236980.6
ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2
|
FAST kinase domains 2 |
| chr20_-_44600810 | 0.34 |
ENST00000322927.2
ENST00000426788.1 |
ZNF335
|
zinc finger protein 335 |
| chr16_+_57769635 | 0.34 |
ENST00000379661.3
ENST00000562592.1 ENST00000566726.1 |
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1 |
| chr4_-_39033963 | 0.34 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr12_-_9913489 | 0.34 |
ENST00000228434.3
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr19_+_41768401 | 0.34 |
ENST00000352456.3
ENST00000595018.1 ENST00000597725.1 |
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr16_+_30484021 | 0.33 |
ENST00000358164.5
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) |
| chr4_+_108910870 | 0.33 |
ENST00000403312.1
ENST00000603302.1 ENST00000309522.3 |
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr2_+_27255806 | 0.33 |
ENST00000238788.9
ENST00000404032.3 |
TMEM214
|
transmembrane protein 214 |
| chr5_-_137374288 | 0.33 |
ENST00000514310.1
|
FAM13B
|
family with sequence similarity 13, member B |
| chr15_+_67835517 | 0.32 |
ENST00000395476.2
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr7_+_56131917 | 0.31 |
ENST00000434526.2
ENST00000275607.9 ENST00000395435.2 ENST00000413952.2 ENST00000342190.6 ENST00000437307.2 ENST00000413756.1 ENST00000451338.1 |
SUMF2
|
sulfatase modifying factor 2 |
| chr11_+_119038897 | 0.31 |
ENST00000454811.1
ENST00000449394.1 |
NLRX1
|
NLR family member X1 |
| chr7_-_56160625 | 0.31 |
ENST00000446428.1
ENST00000432123.1 ENST00000452681.2 ENST00000537360.1 |
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr13_+_49684445 | 0.31 |
ENST00000398316.3
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr8_-_121457332 | 0.30 |
ENST00000518918.1
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr12_-_125398850 | 0.30 |
ENST00000535859.1
ENST00000546271.1 ENST00000540700.1 ENST00000546120.1 |
UBC
|
ubiquitin C |
| chr11_+_66276550 | 0.30 |
ENST00000419755.3
|
CTD-3074O7.11
|
Bardet-Biedl syndrome 1 protein |
| chr7_+_16700806 | 0.29 |
ENST00000446596.1
ENST00000438834.1 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr2_-_114036488 | 0.29 |
ENST00000263335.7
ENST00000397647.3 ENST00000348715.5 ENST00000429538.3 |
PAX8
|
paired box 8 |
| chr14_-_77787198 | 0.29 |
ENST00000261534.4
|
POMT2
|
protein-O-mannosyltransferase 2 |
| chr8_+_145218673 | 0.29 |
ENST00000326134.5
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr11_+_86748998 | 0.28 |
ENST00000525018.1
ENST00000355734.4 |
TMEM135
|
transmembrane protein 135 |
| chr14_-_22005343 | 0.28 |
ENST00000327430.3
|
SALL2
|
spalt-like transcription factor 2 |
| chr15_+_81589254 | 0.27 |
ENST00000394652.2
|
IL16
|
interleukin 16 |
| chr17_-_5026397 | 0.27 |
ENST00000250076.3
|
ZNF232
|
zinc finger protein 232 |
| chr17_+_78152274 | 0.26 |
ENST00000344227.2
ENST00000570421.1 |
CARD14
|
caspase recruitment domain family, member 14 |
| chr17_+_42977062 | 0.26 |
ENST00000410006.2
ENST00000357776.2 ENST00000410027.1 |
CCDC103
|
coiled-coil domain containing 103 |
| chr19_+_39930212 | 0.26 |
ENST00000396843.1
|
AC011500.1
|
Interleukin-like; Uncharacterized protein |
| chr15_-_90892669 | 0.25 |
ENST00000412799.2
|
GABARAPL3
|
GABA(A) receptors associated protein like 3, pseudogene |
| chr1_+_22962948 | 0.24 |
ENST00000374642.3
|
C1QA
|
complement component 1, q subcomponent, A chain |
| chr21_-_46340770 | 0.23 |
ENST00000397854.3
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr22_-_26875345 | 0.23 |
ENST00000398141.1
|
HPS4
|
Hermansky-Pudlak syndrome 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1H2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.6 | GO:0036100 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.8 | 6.1 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.5 | 1.4 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.4 | 3.1 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.4 | 2.5 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.4 | 1.9 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.3 | 1.9 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.3 | 2.8 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.3 | 0.8 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.3 | 1.3 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.3 | 2.4 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.3 | 1.8 | GO:1903281 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.3 | 2.4 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.3 | 2.3 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.3 | 3.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.3 | 0.8 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.3 | 1.8 | GO:0060356 | leucine import(GO:0060356) |
| 0.2 | 0.7 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.2 | 1.9 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.2 | 1.5 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.2 | 2.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.2 | 1.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 1.7 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.1 | 2.8 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 3.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 1.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.9 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 1.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 1.9 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.1 | 1.2 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 2.4 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.1 | 6.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 0.3 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 0.6 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 2.0 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.9 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 | 0.3 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 0.6 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.1 | 0.2 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.1 | 1.2 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 1.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.3 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 1.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.4 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.1 | 0.5 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.3 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.1 | 0.8 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 1.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 0.8 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 2.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.2 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.7 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.9 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 4.5 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 1.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 2.7 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 | 0.2 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 4.2 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.8 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 1.1 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.5 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 1.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 3.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.0 | 8.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.3 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.4 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.8 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 1.8 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.4 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 0.0 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.8 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 1.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 1.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.6 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.3 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.3 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 2.3 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.2 | 2.0 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.2 | 2.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 2.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.6 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 7.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 1.9 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.2 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.1 | 1.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 2.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.6 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.1 | 0.8 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 0.4 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 0.7 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 6.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 3.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.1 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 5.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 3.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.0 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0070702 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.0 | 0.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.4 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.9 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.8 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.3 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 4.1 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 7.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 1.7 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.7 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.7 | GO:0031966 | mitochondrial membrane(GO:0031966) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0097259 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 1.1 | 3.4 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.9 | 2.8 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.8 | 2.3 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.5 | 3.1 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.5 | 4.5 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.4 | 1.3 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.4 | 1.2 | GO:0008124 | phenylalanine 4-monooxygenase activity(GO:0004505) 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.4 | 1.5 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.4 | 1.1 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.3 | 3.8 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.3 | 1.0 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.3 | 1.3 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.3 | 1.6 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.3 | 0.9 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.3 | 5.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 0.9 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.2 | 0.8 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.2 | 0.7 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.2 | 7.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.9 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 0.4 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.1 | 2.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 1.9 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 1.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 2.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.6 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 1.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 2.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.0 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 7.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.6 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.1 | 1.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 7.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.6 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 0.6 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 3.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.3 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0016429 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.2 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 2.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.6 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 5.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.9 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.3 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.0 | GO:0004794 | L-serine ammonia-lyase activity(GO:0003941) L-threonine ammonia-lyase activity(GO:0004794) |
| 0.0 | 0.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 2.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 1.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 2.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 12.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 2.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 3.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 7.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.8 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 3.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 3.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 6.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 1.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 5.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.8 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.1 | 1.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 2.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 5.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.9 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 2.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.7 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |