Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
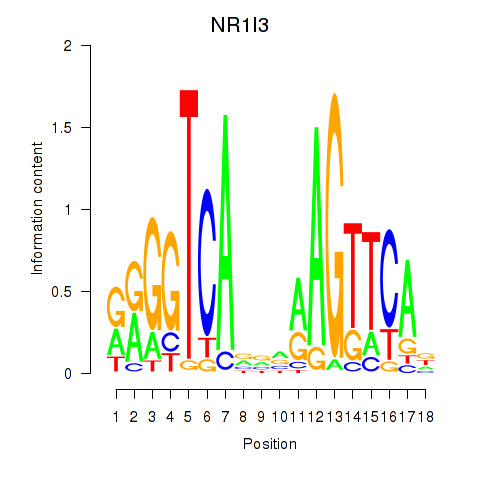
Results for NR1I3
Z-value: 0.43
Transcription factors associated with NR1I3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1I3
|
ENSG00000143257.7 | nuclear receptor subfamily 1 group I member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1I3 | hg19_v2_chr1_-_161207875_161207901 | -0.25 | 2.8e-01 | Click! |
Activity profile of NR1I3 motif
Sorted Z-values of NR1I3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_32582293 | 2.22 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chr19_+_2977444 | 1.25 |
ENST00000246112.4
ENST00000453329.1 ENST00000482627.1 ENST00000452088.1 |
TLE6
|
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
| chr9_-_75567962 | 1.21 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr17_-_26695013 | 1.12 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr17_-_26694979 | 1.08 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr6_-_127780510 | 0.87 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr16_-_16317321 | 0.74 |
ENST00000205557.7
ENST00000575728.1 |
ABCC6
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6 |
| chr16_-_66968265 | 0.68 |
ENST00000567511.1
ENST00000422424.2 |
FAM96B
|
family with sequence similarity 96, member B |
| chr1_-_23886285 | 0.60 |
ENST00000374561.5
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr16_-_66968055 | 0.58 |
ENST00000568572.1
|
FAM96B
|
family with sequence similarity 96, member B |
| chr1_-_32210275 | 0.57 |
ENST00000440175.2
|
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr20_+_62339381 | 0.55 |
ENST00000355969.6
ENST00000357119.4 ENST00000431125.1 ENST00000369967.3 ENST00000328969.5 |
ZGPAT
|
zinc finger, CCCH-type with G patch domain |
| chr17_-_8198636 | 0.54 |
ENST00000577745.1
ENST00000579192.1 ENST00000396278.1 |
SLC25A35
|
solute carrier family 25, member 35 |
| chr12_+_57998400 | 0.53 |
ENST00000548804.1
ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr19_-_46285646 | 0.51 |
ENST00000458663.2
|
DMPK
|
dystrophia myotonica-protein kinase |
| chr20_+_5987890 | 0.48 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr19_-_9968816 | 0.47 |
ENST00000590841.1
|
OLFM2
|
olfactomedin 2 |
| chr1_-_151119087 | 0.44 |
ENST00000341697.3
ENST00000368914.3 |
SEMA6C
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr19_-_7562334 | 0.43 |
ENST00000593942.1
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma |
| chr16_+_577697 | 0.43 |
ENST00000562370.1
ENST00000568988.1 ENST00000219611.2 |
CAPN15
|
calpain 15 |
| chr1_-_155159748 | 0.42 |
ENST00000473363.2
|
RP11-201K10.3
|
RP11-201K10.3 |
| chr1_+_45274154 | 0.42 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr19_-_48894104 | 0.41 |
ENST00000597017.1
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr5_+_139055021 | 0.40 |
ENST00000502716.1
ENST00000503511.1 |
CXXC5
|
CXXC finger protein 5 |
| chr19_+_7562431 | 0.40 |
ENST00000361664.2
|
C19orf45
|
chromosome 19 open reading frame 45 |
| chr5_+_139055055 | 0.39 |
ENST00000511457.1
|
CXXC5
|
CXXC finger protein 5 |
| chrX_-_110655391 | 0.37 |
ENST00000356915.2
ENST00000356220.3 |
DCX
|
doublecortin |
| chr17_-_53499310 | 0.37 |
ENST00000262065.3
|
MMD
|
monocyte to macrophage differentiation-associated |
| chr2_-_228028829 | 0.37 |
ENST00000396625.3
ENST00000329662.7 |
COL4A4
|
collagen, type IV, alpha 4 |
| chr2_+_220462560 | 0.36 |
ENST00000456909.1
ENST00000295641.10 |
STK11IP
|
serine/threonine kinase 11 interacting protein |
| chr5_+_52856456 | 0.36 |
ENST00000296684.5
ENST00000506765.1 |
NDUFS4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase) |
| chr12_+_53693812 | 0.35 |
ENST00000549488.1
|
C12orf10
|
chromosome 12 open reading frame 10 |
| chr3_-_58419537 | 0.33 |
ENST00000474765.1
ENST00000485460.1 ENST00000302746.6 ENST00000383714.4 |
PDHB
|
pyruvate dehydrogenase (lipoamide) beta |
| chr18_-_19284724 | 0.33 |
ENST00000580981.1
ENST00000289119.2 |
ABHD3
|
abhydrolase domain containing 3 |
| chr10_-_73848764 | 0.30 |
ENST00000317376.4
ENST00000412663.1 |
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr19_-_49140692 | 0.30 |
ENST00000222122.5
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr19_-_6604094 | 0.30 |
ENST00000597430.2
|
CD70
|
CD70 molecule |
| chr8_-_109799793 | 0.28 |
ENST00000297459.3
|
TMEM74
|
transmembrane protein 74 |
| chr14_+_77787227 | 0.27 |
ENST00000216465.5
ENST00000361389.4 ENST00000554279.1 ENST00000557639.1 ENST00000349555.3 ENST00000556627.1 ENST00000557053.1 |
GSTZ1
|
glutathione S-transferase zeta 1 |
| chr2_-_241075706 | 0.26 |
ENST00000607357.1
ENST00000307266.3 |
MYEOV2
|
myeloma overexpressed 2 |
| chr7_-_56160625 | 0.23 |
ENST00000446428.1
ENST00000432123.1 ENST00000452681.2 ENST00000537360.1 |
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr9_+_35605274 | 0.23 |
ENST00000336395.5
|
TESK1
|
testis-specific kinase 1 |
| chr2_+_177015950 | 0.22 |
ENST00000306324.3
|
HOXD4
|
homeobox D4 |
| chr1_+_113217043 | 0.22 |
ENST00000413052.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr19_-_49140609 | 0.22 |
ENST00000601104.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr6_+_108487245 | 0.22 |
ENST00000368986.4
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr3_+_52350335 | 0.21 |
ENST00000420323.2
|
DNAH1
|
dynein, axonemal, heavy chain 1 |
| chr15_+_85523671 | 0.20 |
ENST00000310298.4
ENST00000557957.1 |
PDE8A
|
phosphodiesterase 8A |
| chr4_+_70894130 | 0.20 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr2_-_69180012 | 0.17 |
ENST00000481498.1
|
GKN2
|
gastrokine 2 |
| chr11_+_62556596 | 0.17 |
ENST00000526546.1
|
TMEM179B
|
transmembrane protein 179B |
| chr6_-_31509714 | 0.16 |
ENST00000456662.1
ENST00000431908.1 ENST00000456976.1 ENST00000428450.1 ENST00000453105.2 ENST00000418897.1 ENST00000415382.2 ENST00000449074.2 ENST00000419020.1 ENST00000428098.1 |
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr1_-_205290865 | 0.15 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chr10_+_47658234 | 0.15 |
ENST00000447511.2
ENST00000537271.1 |
ANTXRL
|
anthrax toxin receptor-like |
| chr11_-_82611448 | 0.15 |
ENST00000393399.2
ENST00000313010.3 |
PRCP
|
prolylcarboxypeptidase (angiotensinase C) |
| chr16_-_4465886 | 0.15 |
ENST00000539968.1
|
CORO7
|
coronin 7 |
| chr18_-_33077556 | 0.14 |
ENST00000589273.1
ENST00000586489.1 |
INO80C
|
INO80 complex subunit C |
| chr11_-_59950519 | 0.14 |
ENST00000528851.1
|
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr22_+_42372764 | 0.14 |
ENST00000396426.3
ENST00000406029.1 |
SEPT3
|
septin 3 |
| chr6_-_31509506 | 0.14 |
ENST00000449757.1
|
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr20_-_62339190 | 0.14 |
ENST00000324228.2
ENST00000609142.1 |
ARFRP1
|
ADP-ribosylation factor related protein 1 |
| chr13_-_114107839 | 0.14 |
ENST00000375418.3
|
ADPRHL1
|
ADP-ribosylhydrolase like 1 |
| chr11_-_83878041 | 0.14 |
ENST00000398299.1
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr3_-_50396978 | 0.14 |
ENST00000266025.3
|
TMEM115
|
transmembrane protein 115 |
| chr19_-_46296011 | 0.13 |
ENST00000377735.3
ENST00000270223.6 |
DMWD
|
dystrophia myotonica, WD repeat containing |
| chr14_-_22005062 | 0.13 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr18_-_19283649 | 0.13 |
ENST00000584464.1
ENST00000578270.1 |
ABHD3
|
abhydrolase domain containing 3 |
| chr1_+_32930647 | 0.11 |
ENST00000609129.1
|
ZBTB8B
|
zinc finger and BTB domain containing 8B |
| chr17_-_62502399 | 0.11 |
ENST00000450599.2
ENST00000585060.1 |
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr19_+_58920102 | 0.11 |
ENST00000599238.1
ENST00000322834.7 |
ZNF584
|
zinc finger protein 584 |
| chr3_+_122103014 | 0.11 |
ENST00000232125.5
ENST00000477892.1 ENST00000469967.1 |
FAM162A
|
family with sequence similarity 162, member A |
| chr4_-_170897045 | 0.11 |
ENST00000508313.1
|
RP11-205M3.3
|
RP11-205M3.3 |
| chr2_-_69180083 | 0.10 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chr17_-_53499218 | 0.10 |
ENST00000571578.1
|
MMD
|
monocyte to macrophage differentiation-associated |
| chr1_-_9086404 | 0.10 |
ENST00000400906.1
|
SLC2A7
|
solute carrier family 2 (facilitated glucose transporter), member 7 |
| chr13_+_24883703 | 0.10 |
ENST00000332018.4
|
C1QTNF9
|
C1q and tumor necrosis factor related protein 9 |
| chr16_-_75018968 | 0.09 |
ENST00000262144.6
|
WDR59
|
WD repeat domain 59 |
| chr9_-_91793675 | 0.09 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr1_-_149900122 | 0.09 |
ENST00000271628.8
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa |
| chr1_-_33336414 | 0.09 |
ENST00000373471.3
ENST00000609187.1 |
FNDC5
|
fibronectin type III domain containing 5 |
| chr16_+_67280799 | 0.09 |
ENST00000566345.2
|
SLC9A5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr12_-_54779511 | 0.09 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr10_-_50396407 | 0.08 |
ENST00000374153.2
ENST00000374151.3 |
C10orf128
|
chromosome 10 open reading frame 128 |
| chr1_-_20126365 | 0.08 |
ENST00000294543.6
ENST00000375122.2 |
TMCO4
|
transmembrane and coiled-coil domains 4 |
| chrX_+_110924346 | 0.08 |
ENST00000371979.3
ENST00000251943.4 ENST00000486353.1 ENST00000394780.3 ENST00000495283.1 |
ALG13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr22_+_39493207 | 0.08 |
ENST00000348946.4
ENST00000442487.3 ENST00000421988.2 |
APOBEC3H
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
| chrX_-_39186610 | 0.08 |
ENST00000429281.1
ENST00000448597.1 |
RP11-265P11.2
|
RP11-265P11.2 |
| chr9_-_35361262 | 0.07 |
ENST00000599954.1
|
AL160274.1
|
HCG17281; PRO0038; Uncharacterized protein |
| chr14_-_77787198 | 0.07 |
ENST00000261534.4
|
POMT2
|
protein-O-mannosyltransferase 2 |
| chr11_+_63137251 | 0.07 |
ENST00000310969.4
ENST00000279178.3 |
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9 |
| chr3_-_116164306 | 0.07 |
ENST00000490035.2
|
LSAMP
|
limbic system-associated membrane protein |
| chr8_-_52721975 | 0.06 |
ENST00000356297.4
ENST00000543296.1 |
PXDNL
|
peroxidasin homolog (Drosophila)-like |
| chr1_+_59250815 | 0.06 |
ENST00000544621.1
ENST00000419531.2 |
RP4-794H19.2
|
long intergenic non-protein coding RNA 1135 |
| chr17_-_33390667 | 0.06 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr19_+_58919992 | 0.06 |
ENST00000306910.4
ENST00000598901.1 ENST00000593920.1 ENST00000596281.1 |
ZNF584
|
zinc finger protein 584 |
| chr20_-_44007014 | 0.06 |
ENST00000372726.3
ENST00000537995.1 |
TP53TG5
|
TP53 target 5 |
| chr19_+_676385 | 0.06 |
ENST00000166139.4
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein) |
| chr1_+_43803475 | 0.06 |
ENST00000372470.3
ENST00000413998.2 |
MPL
|
myeloproliferative leukemia virus oncogene |
| chr6_-_43496605 | 0.06 |
ENST00000455285.2
|
XPO5
|
exportin 5 |
| chr11_-_104480019 | 0.05 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr12_-_75601778 | 0.05 |
ENST00000550433.1
ENST00000548513.1 |
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr5_+_156712372 | 0.05 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chrX_+_118602363 | 0.05 |
ENST00000317881.8
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr14_+_100070869 | 0.05 |
ENST00000502101.2
|
RP11-543C4.1
|
RP11-543C4.1 |
| chr12_+_49208234 | 0.05 |
ENST00000540990.1
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr20_-_62339315 | 0.05 |
ENST00000440854.1
ENST00000607873.1 |
ARFRP1
|
ADP-ribosylation factor related protein 1 |
| chr7_-_56160666 | 0.05 |
ENST00000297373.2
|
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr17_+_26833250 | 0.05 |
ENST00000577936.1
ENST00000579795.1 |
FOXN1
|
forkhead box N1 |
| chr11_-_83393303 | 0.04 |
ENST00000398304.1
ENST00000420775.2 |
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr6_-_34113856 | 0.04 |
ENST00000538487.2
|
GRM4
|
glutamate receptor, metabotropic 4 |
| chr1_-_27682962 | 0.04 |
ENST00000486046.1
|
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr4_+_37245799 | 0.04 |
ENST00000309447.5
|
KIAA1239
|
KIAA1239 |
| chr2_-_236692031 | 0.04 |
ENST00000440101.1
|
AC064874.1
|
Uncharacterized protein |
| chr12_-_108991778 | 0.04 |
ENST00000549447.1
|
TMEM119
|
transmembrane protein 119 |
| chr1_+_160097462 | 0.03 |
ENST00000447527.1
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr22_+_39493268 | 0.03 |
ENST00000401756.1
|
APOBEC3H
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
| chr8_+_11351876 | 0.03 |
ENST00000529894.1
|
BLK
|
B lymphoid tyrosine kinase |
| chr9_+_37486005 | 0.03 |
ENST00000377792.3
|
POLR1E
|
polymerase (RNA) I polypeptide E, 53kDa |
| chr3_-_116163830 | 0.02 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr1_+_26036093 | 0.02 |
ENST00000374329.1
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1 |
| chr17_-_34270697 | 0.02 |
ENST00000585556.1
|
LYZL6
|
lysozyme-like 6 |
| chr11_+_105480740 | 0.01 |
ENST00000393125.2
|
GRIA4
|
glutamate receptor, ionotropic, AMPA 4 |
| chr17_-_34270668 | 0.01 |
ENST00000293274.4
|
LYZL6
|
lysozyme-like 6 |
| chr10_+_47657581 | 0.01 |
ENST00000441923.2
|
ANTXRL
|
anthrax toxin receptor-like |
| chr17_-_3301704 | 0.01 |
ENST00000322608.2
|
OR1E1
|
olfactory receptor, family 1, subfamily E, member 1 |
| chr12_-_108991812 | 0.01 |
ENST00000392806.3
ENST00000547567.1 |
TMEM119
|
transmembrane protein 119 |
| chr10_+_88414338 | 0.00 |
ENST00000241891.5
ENST00000443292.1 |
OPN4
|
opsin 4 |
| chr13_-_95131923 | 0.00 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr9_-_80646374 | 0.00 |
ENST00000286548.4
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1I3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.2 | 1.2 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 1.2 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 0.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.5 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.1 | 0.4 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 1.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 1.1 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.5 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.3 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.3 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.4 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.2 | GO:0002353 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.0 | 0.2 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.6 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.4 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 0.1 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.0 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) regulation of positive thymic T cell selection(GO:1902232) |
| 0.0 | 0.1 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.0 | GO:1903280 | negative regulation of calcium:sodium antiporter activity(GO:1903280) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.2 | 1.3 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.4 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.3 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.1 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.4 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.1 | 1.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.3 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.4 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.1 | 0.5 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 2.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.7 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 1.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.1 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.2 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |