Project
SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
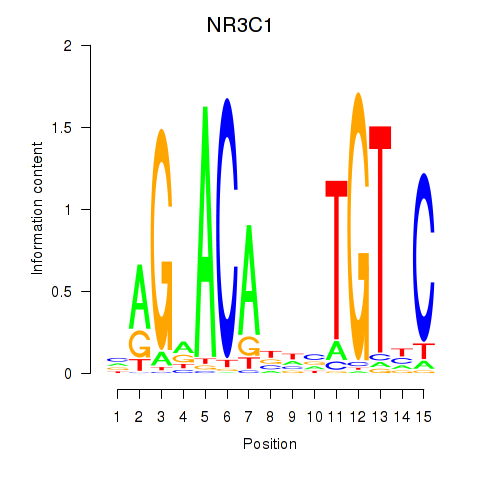
Results for NR3C1
Z-value: 1.75
Transcription factors associated with NR3C1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR3C1
|
ENSG00000113580.10 | nuclear receptor subfamily 3 group C member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR3C1 | hg19_v2_chr5_-_142782862_142782894 | -0.59 | 5.7e-03 | Click! |
Activity profile of NR3C1 motif
Sorted Z-values of NR3C1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_56659687 | 8.92 |
ENST00000568293.1
ENST00000330439.6 |
MT1E
|
metallothionein 1E |
| chr18_+_21464737 | 6.80 |
ENST00000586751.1
|
LAMA3
|
laminin, alpha 3 |
| chr16_+_68679193 | 6.46 |
ENST00000581171.1
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental) |
| chr2_-_113594279 | 6.33 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr3_+_122044084 | 5.79 |
ENST00000264474.3
ENST00000479204.1 |
CSTA
|
cystatin A (stefin A) |
| chr1_-_209979465 | 5.73 |
ENST00000542854.1
|
IRF6
|
interferon regulatory factor 6 |
| chr2_+_208104497 | 5.55 |
ENST00000430494.1
|
AC007879.7
|
AC007879.7 |
| chr2_+_233925064 | 5.43 |
ENST00000359570.5
ENST00000538935.1 |
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr1_+_35247859 | 5.14 |
ENST00000373362.3
|
GJB3
|
gap junction protein, beta 3, 31kDa |
| chr16_+_68678739 | 5.11 |
ENST00000264012.4
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental) |
| chr11_+_57365150 | 4.98 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr10_-_105845674 | 4.96 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr8_-_49834299 | 4.77 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr22_+_24322322 | 4.74 |
ENST00000215780.5
ENST00000402588.3 |
GSTT2
|
glutathione S-transferase theta 2 |
| chr8_-_49833978 | 4.62 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr1_-_27816556 | 4.54 |
ENST00000536657.1
|
WASF2
|
WAS protein family, member 2 |
| chr16_+_56716336 | 4.46 |
ENST00000394485.4
ENST00000562939.1 |
MT1X
|
metallothionein 1X |
| chr14_-_23623577 | 4.39 |
ENST00000422941.2
ENST00000453702.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr1_-_153013588 | 4.31 |
ENST00000360379.3
|
SPRR2D
|
small proline-rich protein 2D |
| chr10_-_105845536 | 4.16 |
ENST00000393211.3
|
COL17A1
|
collagen, type XVII, alpha 1 |
| chr5_-_149792295 | 4.09 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr8_-_125740514 | 4.05 |
ENST00000325064.5
ENST00000518547.1 |
MTSS1
|
metastasis suppressor 1 |
| chr17_-_5487768 | 3.91 |
ENST00000269280.4
ENST00000345221.3 ENST00000262467.5 |
NLRP1
|
NLR family, pyrin domain containing 1 |
| chr1_+_6511651 | 3.85 |
ENST00000434576.1
|
ESPN
|
espin |
| chr11_-_102668879 | 3.78 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr19_-_51456344 | 3.74 |
ENST00000336334.3
ENST00000593428.1 |
KLK5
|
kallikrein-related peptidase 5 |
| chr7_-_20256965 | 3.72 |
ENST00000400331.5
ENST00000332878.4 |
MACC1
|
metastasis associated in colon cancer 1 |
| chr11_+_5711010 | 3.71 |
ENST00000454828.1
|
TRIM22
|
tripartite motif containing 22 |
| chr16_+_56642489 | 3.52 |
ENST00000561491.1
|
MT2A
|
metallothionein 2A |
| chr12_-_28125638 | 3.52 |
ENST00000545234.1
|
PTHLH
|
parathyroid hormone-like hormone |
| chr8_-_125740730 | 3.46 |
ENST00000354184.4
|
MTSS1
|
metastasis suppressor 1 |
| chr13_+_78109804 | 3.42 |
ENST00000535157.1
|
SCEL
|
sciellin |
| chr12_+_76653611 | 3.39 |
ENST00000550380.1
|
RP11-54A9.1
|
RP11-54A9.1 |
| chr17_-_39661849 | 3.36 |
ENST00000246635.3
ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13
|
keratin 13 |
| chr5_+_96212185 | 3.35 |
ENST00000379904.4
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr6_+_41604747 | 3.30 |
ENST00000419164.1
ENST00000373051.2 |
MDFI
|
MyoD family inhibitor |
| chr1_-_209792111 | 3.28 |
ENST00000455193.1
|
LAMB3
|
laminin, beta 3 |
| chr11_-_12030681 | 3.23 |
ENST00000529338.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr5_+_49961727 | 3.22 |
ENST00000505697.2
ENST00000503750.2 ENST00000514342.2 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr6_+_18387570 | 3.16 |
ENST00000259939.3
|
RNF144B
|
ring finger protein 144B |
| chr2_+_208104351 | 3.16 |
ENST00000440326.1
|
AC007879.7
|
AC007879.7 |
| chr19_-_51456321 | 3.13 |
ENST00000391809.2
|
KLK5
|
kallikrein-related peptidase 5 |
| chr19_+_53868940 | 3.13 |
ENST00000474037.1
ENST00000491101.1 ENST00000467003.1 ENST00000475179.1 ENST00000593918.1 |
ZNF525
|
zinc finger protein 525 |
| chr15_-_72490114 | 3.09 |
ENST00000309731.7
|
GRAMD2
|
GRAM domain containing 2 |
| chr6_+_41604620 | 3.07 |
ENST00000432027.1
|
MDFI
|
MyoD family inhibitor |
| chr17_+_7344057 | 3.07 |
ENST00000575398.1
ENST00000575082.1 |
FGF11
|
fibroblast growth factor 11 |
| chr12_+_76653682 | 3.05 |
ENST00000553247.1
|
RP11-54A9.1
|
RP11-54A9.1 |
| chr1_-_17304771 | 3.02 |
ENST00000375534.3
|
MFAP2
|
microfibrillar-associated protein 2 |
| chr19_-_57988871 | 2.93 |
ENST00000596831.1
ENST00000601768.1 ENST00000356584.3 ENST00000600175.1 ENST00000425074.3 ENST00000343280.4 ENST00000427512.2 |
AC004076.9
ZNF772
|
Uncharacterized protein zinc finger protein 772 |
| chr6_+_29795595 | 2.92 |
ENST00000360323.6
ENST00000376818.3 ENST00000376815.3 |
HLA-G
|
major histocompatibility complex, class I, G |
| chr13_+_78109884 | 2.91 |
ENST00000377246.3
ENST00000349847.3 |
SCEL
|
sciellin |
| chr18_-_43652211 | 2.89 |
ENST00000589328.1
ENST00000409746.5 |
PSTPIP2
|
proline-serine-threonine phosphatase interacting protein 2 |
| chr19_-_36822551 | 2.85 |
ENST00000591372.1
|
LINC00665
|
long intergenic non-protein coding RNA 665 |
| chr6_-_47010061 | 2.84 |
ENST00000371253.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr11_+_18287721 | 2.82 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr18_+_61442629 | 2.81 |
ENST00000398019.2
ENST00000540675.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr4_-_153601136 | 2.80 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr17_-_71410794 | 2.76 |
ENST00000424778.1
|
SDK2
|
sidekick cell adhesion molecule 2 |
| chr14_+_94640633 | 2.75 |
ENST00000304338.3
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4 |
| chr1_-_182360918 | 2.75 |
ENST00000339526.4
|
GLUL
|
glutamate-ammonia ligase |
| chr1_-_182360498 | 2.66 |
ENST00000417584.2
|
GLUL
|
glutamate-ammonia ligase |
| chr3_-_42845922 | 2.64 |
ENST00000452906.2
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A |
| chr7_+_40174565 | 2.62 |
ENST00000309930.5
ENST00000401647.2 ENST00000335693.4 ENST00000413931.1 ENST00000416370.1 ENST00000540834.1 |
C7orf10
|
succinylCoA:glutarate-CoA transferase |
| chr11_-_118135160 | 2.62 |
ENST00000438295.2
|
MPZL2
|
myelin protein zero-like 2 |
| chrX_+_135618258 | 2.59 |
ENST00000440515.1
ENST00000456412.1 |
VGLL1
|
vestigial like 1 (Drosophila) |
| chr17_+_38333263 | 2.59 |
ENST00000456989.2
ENST00000543876.1 ENST00000544503.1 ENST00000264644.6 ENST00000538884.1 |
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr12_+_66582919 | 2.58 |
ENST00000545837.1
ENST00000457197.2 |
IRAK3
|
interleukin-1 receptor-associated kinase 3 |
| chr11_+_18287801 | 2.56 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr20_+_12989596 | 2.53 |
ENST00000434210.1
ENST00000399002.2 |
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr13_-_38172863 | 2.44 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chrX_+_152082969 | 2.44 |
ENST00000535861.1
ENST00000539731.1 ENST00000449285.2 ENST00000318504.7 ENST00000324823.6 ENST00000370268.4 ENST00000370270.2 |
ZNF185
|
zinc finger protein 185 (LIM domain) |
| chr12_-_15114492 | 2.34 |
ENST00000541546.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr10_-_4720333 | 2.30 |
ENST00000430998.2
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr8_-_125577940 | 2.29 |
ENST00000519168.1
ENST00000395508.2 |
MTSS1
|
metastasis suppressor 1 |
| chr14_+_94640671 | 2.28 |
ENST00000328839.3
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4 |
| chr12_-_15114191 | 2.27 |
ENST00000541380.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr12_-_95009837 | 2.27 |
ENST00000551457.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr11_+_34663913 | 2.25 |
ENST00000532302.1
|
EHF
|
ets homologous factor |
| chr22_-_24303340 | 2.24 |
ENST00000404172.3
ENST00000290765.4 |
GSTT2B
|
glutathione S-transferase theta 2B (gene/pseudogene) |
| chr3_+_38035610 | 2.23 |
ENST00000465644.1
|
VILL
|
villin-like |
| chr19_+_41222998 | 2.22 |
ENST00000263370.2
|
ITPKC
|
inositol-trisphosphate 3-kinase C |
| chr19_-_9811347 | 2.21 |
ENST00000585964.1
ENST00000457674.2 ENST00000590544.1 |
ZNF812
|
zinc finger protein 812 |
| chr3_-_42846021 | 2.21 |
ENST00000321331.7
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A |
| chr17_-_39661947 | 2.20 |
ENST00000590425.1
|
KRT13
|
keratin 13 |
| chr19_+_53073526 | 2.19 |
ENST00000596514.1
ENST00000391785.3 ENST00000301093.2 |
ZNF701
|
zinc finger protein 701 |
| chr11_+_101983176 | 2.18 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr19_-_20748614 | 2.18 |
ENST00000596797.1
|
ZNF737
|
zinc finger protein 737 |
| chrX_+_56259316 | 2.17 |
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8 |
| chr19_+_56905024 | 2.16 |
ENST00000591172.1
ENST00000589888.1 ENST00000587979.1 ENST00000585659.1 ENST00000593109.1 |
ZNF582-AS1
|
ZNF582 antisense RNA 1 (head to head) |
| chr14_+_61654271 | 2.16 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr13_-_108867846 | 2.16 |
ENST00000442234.1
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr3_+_195447738 | 2.15 |
ENST00000447234.2
ENST00000320736.6 ENST00000436408.1 |
MUC20
|
mucin 20, cell surface associated |
| chr11_-_57194550 | 2.13 |
ENST00000528187.1
ENST00000524863.1 ENST00000533051.1 ENST00000529494.1 ENST00000395124.1 ENST00000533524.1 ENST00000533245.1 ENST00000530316.1 |
SLC43A3
|
solute carrier family 43, member 3 |
| chr4_+_87928353 | 2.13 |
ENST00000511722.1
|
AFF1
|
AF4/FMR2 family, member 1 |
| chrX_-_30885319 | 2.11 |
ENST00000378933.1
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chrX_-_30877837 | 2.11 |
ENST00000378930.3
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr11_+_34664014 | 2.11 |
ENST00000527935.1
|
EHF
|
ets homologous factor |
| chr7_-_121784285 | 2.10 |
ENST00000417368.2
|
AASS
|
aminoadipate-semialdehyde synthase |
| chr5_+_67576062 | 2.10 |
ENST00000523807.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr2_-_74875432 | 2.10 |
ENST00000536235.1
ENST00000421985.1 |
M1AP
|
meiosis 1 associated protein |
| chr3_-_121379739 | 2.09 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr7_+_143318020 | 2.08 |
ENST00000444908.2
ENST00000518791.1 ENST00000411497.2 |
FAM115C
|
family with sequence similarity 115, member C |
| chr17_+_74380683 | 2.08 |
ENST00000592299.1
ENST00000590959.1 ENST00000323374.4 |
SPHK1
|
sphingosine kinase 1 |
| chr1_+_152956549 | 2.07 |
ENST00000307122.2
|
SPRR1A
|
small proline-rich protein 1A |
| chr2_-_214016314 | 2.04 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr11_-_111783595 | 2.03 |
ENST00000528628.1
|
CRYAB
|
crystallin, alpha B |
| chr9_+_112887772 | 2.02 |
ENST00000259318.7
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr17_+_74381343 | 2.02 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr13_+_21714653 | 1.95 |
ENST00000382533.4
|
SAP18
|
Sin3A-associated protein, 18kDa |
| chr16_+_56642041 | 1.95 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A |
| chr13_-_36920872 | 1.94 |
ENST00000451493.1
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr11_-_7698453 | 1.94 |
ENST00000524608.1
|
CYB5R2
|
cytochrome b5 reductase 2 |
| chr4_+_75311019 | 1.93 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr1_+_104104379 | 1.92 |
ENST00000435302.1
|
AMY2B
|
amylase, alpha 2B (pancreatic) |
| chr22_-_43567750 | 1.90 |
ENST00000494035.1
|
TTLL12
|
tubulin tyrosine ligase-like family, member 12 |
| chr18_+_55816546 | 1.90 |
ENST00000435432.2
ENST00000357895.5 ENST00000586263.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr2_+_66666432 | 1.89 |
ENST00000495021.2
|
MEIS1
|
Meis homeobox 1 |
| chr14_-_61748550 | 1.88 |
ENST00000555868.1
|
TMEM30B
|
transmembrane protein 30B |
| chr8_+_91013577 | 1.88 |
ENST00000220764.2
|
DECR1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr5_-_1801408 | 1.87 |
ENST00000505818.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr3_-_182833863 | 1.87 |
ENST00000492597.1
|
MCCC1
|
methylcrotonoyl-CoA carboxylase 1 (alpha) |
| chr5_-_42811986 | 1.87 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr3_-_16554403 | 1.87 |
ENST00000449415.1
ENST00000441460.1 |
RFTN1
|
raftlin, lipid raft linker 1 |
| chr10_-_4720301 | 1.85 |
ENST00000449712.1
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr12_+_128399965 | 1.84 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr15_-_81616446 | 1.83 |
ENST00000302824.6
|
STARD5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr22_+_21213771 | 1.83 |
ENST00000439214.1
|
SNAP29
|
synaptosomal-associated protein, 29kDa |
| chr17_+_79031415 | 1.82 |
ENST00000572073.1
ENST00000573677.1 |
BAIAP2
|
BAI1-associated protein 2 |
| chr15_-_79237433 | 1.82 |
ENST00000220166.5
|
CTSH
|
cathepsin H |
| chr11_+_5710919 | 1.80 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr1_-_27816641 | 1.79 |
ENST00000430629.2
|
WASF2
|
WAS protein family, member 2 |
| chr20_-_20693131 | 1.79 |
ENST00000202677.7
|
RALGAPA2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr6_+_144665237 | 1.75 |
ENST00000421035.2
|
UTRN
|
utrophin |
| chr5_-_39219641 | 1.75 |
ENST00000509072.1
ENST00000504542.1 ENST00000505428.1 ENST00000506557.1 |
FYB
|
FYN binding protein |
| chr1_+_145293371 | 1.74 |
ENST00000342960.5
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr11_+_32851487 | 1.73 |
ENST00000257836.3
|
PRRG4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr12_-_95010147 | 1.71 |
ENST00000548918.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr4_+_89444961 | 1.71 |
ENST00000513325.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr15_-_42783303 | 1.69 |
ENST00000565380.1
ENST00000564754.1 |
ZNF106
|
zinc finger protein 106 |
| chr20_-_43977055 | 1.68 |
ENST00000372733.3
ENST00000537976.1 |
SDC4
|
syndecan 4 |
| chr10_-_28571015 | 1.66 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr8_-_42396632 | 1.66 |
ENST00000520179.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr1_-_153066998 | 1.65 |
ENST00000368750.3
|
SPRR2E
|
small proline-rich protein 2E |
| chr14_-_21493649 | 1.65 |
ENST00000553442.1
ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2
|
NDRG family member 2 |
| chr6_-_150217195 | 1.65 |
ENST00000532335.1
|
RAET1E
|
retinoic acid early transcript 1E |
| chr20_-_45985172 | 1.65 |
ENST00000536340.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr19_+_57874835 | 1.64 |
ENST00000543226.1
ENST00000596755.1 ENST00000282282.3 ENST00000597658.1 |
TRAPPC2P1
ZNF547
AC003002.4
|
trafficking protein particle complex 2 pseudogene 1 zinc finger protein 547 Uncharacterized protein |
| chr15_+_31658349 | 1.64 |
ENST00000558844.1
|
KLF13
|
Kruppel-like factor 13 |
| chr1_+_144173162 | 1.64 |
ENST00000356801.6
|
NBPF8
|
neuroblastoma breakpoint family, member 8 |
| chr20_+_35201993 | 1.64 |
ENST00000373872.4
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr4_+_75310851 | 1.64 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr1_-_207119738 | 1.62 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr1_+_44115814 | 1.62 |
ENST00000372396.3
|
KDM4A
|
lysine (K)-specific demethylase 4A |
| chr19_-_53662257 | 1.61 |
ENST00000599096.1
ENST00000334197.7 ENST00000597183.1 ENST00000601804.1 ENST00000601469.2 ENST00000452676.2 |
ZNF347
|
zinc finger protein 347 |
| chrX_-_51239425 | 1.61 |
ENST00000375992.3
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr18_+_34124507 | 1.61 |
ENST00000591635.1
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr20_-_45280091 | 1.60 |
ENST00000396360.1
ENST00000435032.1 ENST00000413164.2 ENST00000372121.1 ENST00000339636.3 |
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 |
| chr1_+_151512775 | 1.60 |
ENST00000368849.3
ENST00000392712.3 ENST00000353024.3 ENST00000368848.2 ENST00000538902.1 |
TUFT1
|
tuftelin 1 |
| chr16_+_82068830 | 1.60 |
ENST00000199936.4
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr14_-_21493884 | 1.60 |
ENST00000556974.1
ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2
|
NDRG family member 2 |
| chr6_+_116575329 | 1.59 |
ENST00000430252.2
ENST00000540275.1 ENST00000448740.2 |
DSE
RP3-486I3.7
|
dermatan sulfate epimerase RP3-486I3.7 |
| chr20_+_12989822 | 1.59 |
ENST00000378194.4
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr19_+_53970970 | 1.58 |
ENST00000468450.1
ENST00000396403.4 ENST00000490956.1 ENST00000396421.4 |
ZNF813
|
zinc finger protein 813 |
| chr15_+_42694573 | 1.58 |
ENST00000397200.4
ENST00000569827.1 |
CAPN3
|
calpain 3, (p94) |
| chr9_-_107690420 | 1.57 |
ENST00000423487.2
ENST00000374733.1 ENST00000374736.3 |
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr6_-_30043539 | 1.56 |
ENST00000376751.3
ENST00000244360.6 |
RNF39
|
ring finger protein 39 |
| chr17_-_73975198 | 1.56 |
ENST00000301608.4
ENST00000588176.1 |
ACOX1
|
acyl-CoA oxidase 1, palmitoyl |
| chr17_+_7461580 | 1.56 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr3_-_75834670 | 1.54 |
ENST00000400845.3
ENST00000477374.1 ENST00000478296.1 |
ZNF717
|
zinc finger protein 717 |
| chr2_-_31637560 | 1.54 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr18_+_43753500 | 1.53 |
ENST00000587591.1
ENST00000588730.1 |
C18orf25
|
chromosome 18 open reading frame 25 |
| chr2_+_170440902 | 1.53 |
ENST00000448752.2
ENST00000418888.1 ENST00000414307.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr3_+_9944303 | 1.51 |
ENST00000421412.1
ENST00000295980.3 |
IL17RE
|
interleukin 17 receptor E |
| chr12_-_48164812 | 1.51 |
ENST00000549151.1
ENST00000548919.1 |
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr15_+_31508174 | 1.49 |
ENST00000559292.2
ENST00000557928.1 |
RP11-16E12.1
|
RP11-16E12.1 |
| chr1_+_117963209 | 1.47 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr12_+_51318513 | 1.47 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr19_+_11877838 | 1.47 |
ENST00000357901.4
ENST00000454339.2 |
ZNF441
|
zinc finger protein 441 |
| chr18_+_47087055 | 1.46 |
ENST00000577628.1
|
LIPG
|
lipase, endothelial |
| chr4_-_68995571 | 1.46 |
ENST00000356291.2
|
TMPRSS11F
|
transmembrane protease, serine 11F |
| chr16_-_4852616 | 1.45 |
ENST00000591392.1
ENST00000587711.1 |
ROGDI
|
rogdi homolog (Drosophila) |
| chrX_+_64887512 | 1.45 |
ENST00000360270.5
|
MSN
|
moesin |
| chr3_-_18480260 | 1.45 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr6_-_91296602 | 1.45 |
ENST00000369325.3
ENST00000369327.3 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr7_-_130353553 | 1.44 |
ENST00000330992.7
ENST00000445977.2 |
COPG2
|
coatomer protein complex, subunit gamma 2 |
| chr9_+_135937365 | 1.44 |
ENST00000372080.4
ENST00000351304.7 |
CEL
|
carboxyl ester lipase |
| chr6_-_138833630 | 1.44 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr11_-_104916034 | 1.42 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr3_+_197687071 | 1.41 |
ENST00000482695.1
ENST00000330198.4 ENST00000419117.1 ENST00000420910.2 ENST00000332636.5 |
LMLN
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr18_+_47087390 | 1.40 |
ENST00000583083.1
|
LIPG
|
lipase, endothelial |
| chr1_+_19967014 | 1.40 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chrX_+_2609356 | 1.39 |
ENST00000381180.3
ENST00000449611.1 |
CD99
|
CD99 molecule |
| chr11_+_65851443 | 1.39 |
ENST00000533756.1
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr10_-_28623368 | 1.38 |
ENST00000441595.2
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr17_+_7462031 | 1.38 |
ENST00000380535.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr1_+_109234907 | 1.38 |
ENST00000370025.4
ENST00000370022.5 ENST00000370021.1 |
PRPF38B
|
pre-mRNA processing factor 38B |
| chr20_+_60813535 | 1.38 |
ENST00000358053.2
ENST00000313733.3 ENST00000439951.2 |
OSBPL2
|
oxysterol binding protein-like 2 |
| chr1_+_158969752 | 1.37 |
ENST00000566111.1
|
IFI16
|
interferon, gamma-inducible protein 16 |
| chr19_-_41934635 | 1.37 |
ENST00000321702.2
|
B3GNT8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
| chrX_-_80457385 | 1.37 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR3C1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.4 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 2.9 | 11.6 | GO:0051796 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 1.4 | 9.8 | GO:0030035 | microspike assembly(GO:0030035) |
| 1.4 | 4.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 1.4 | 5.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 1.3 | 3.9 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 1.2 | 9.9 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 1.1 | 6.9 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 1.1 | 5.5 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.9 | 0.9 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.9 | 6.9 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.8 | 4.1 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.8 | 1.5 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.7 | 2.2 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.7 | 2.9 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.7 | 2.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.7 | 4.6 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.6 | 1.8 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.6 | 2.8 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.5 | 5.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.5 | 19.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.5 | 2.6 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.5 | 6.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.5 | 1.9 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.5 | 1.4 | GO:0050904 | diapedesis(GO:0050904) |
| 0.5 | 1.8 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.4 | 2.7 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.4 | 3.0 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.4 | 2.4 | GO:0032571 | response to vitamin K(GO:0032571) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.4 | 2.4 | GO:1901909 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.4 | 3.2 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.4 | 2.4 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.4 | 1.2 | GO:0051714 | positive regulation of cytolysis in other organism(GO:0051714) |
| 0.4 | 1.2 | GO:2000412 | positive regulation of thymocyte migration(GO:2000412) |
| 0.4 | 1.8 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.4 | 1.1 | GO:1903452 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.4 | 3.2 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.3 | 2.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.3 | 3.1 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.3 | 6.1 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.3 | 1.7 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.3 | 1.6 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.3 | 1.6 | GO:0032489 | aminophospholipid transport(GO:0015917) regulation of Cdc42 protein signal transduction(GO:0032489) regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.3 | 9.4 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.3 | 0.9 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.3 | 1.2 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.3 | 2.4 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.3 | 3.0 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.3 | 1.5 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.3 | 5.1 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.3 | 1.7 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.3 | 1.7 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.3 | 0.9 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.3 | 1.4 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.3 | 1.1 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.3 | 2.3 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.3 | 1.7 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.3 | 1.4 | GO:0035936 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.3 | 1.4 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.3 | 0.3 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.3 | 1.9 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.3 | 1.4 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.3 | 1.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.3 | 1.9 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.3 | 1.6 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.3 | 6.4 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.3 | 2.1 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.3 | 1.3 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.3 | 0.8 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.3 | 3.8 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.3 | 1.8 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.2 | 3.2 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.2 | 8.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 1.2 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.2 | 1.0 | GO:0033594 | response to hydroxyisoflavone(GO:0033594) activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.2 | 1.6 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.2 | 1.4 | GO:1900114 | positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.2 | 1.6 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.2 | 1.6 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.2 | 0.7 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.2 | 1.1 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.2 | 2.2 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.2 | 0.9 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.2 | 0.6 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.2 | 1.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.2 | 0.4 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.2 | 1.5 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.2 | 1.9 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.2 | 1.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 1.4 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.2 | 2.8 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 1.0 | GO:0006726 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.2 | 5.0 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 0.6 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.2 | 1.3 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 1.9 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.2 | 1.5 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 4.8 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.2 | 0.5 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.2 | 0.5 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.2 | 11.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 2.2 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.2 | 5.8 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.1 | 0.6 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.1 | 1.2 | GO:1901297 | arterial endothelial cell fate commitment(GO:0060844) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 8.9 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.1 | 2.4 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.6 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.6 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.1 | 0.9 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 0.8 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 1.1 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 0.5 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.9 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 | 0.4 | GO:0034130 | toll-like receptor 1 signaling pathway(GO:0034130) |
| 0.1 | 0.5 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.1 | 1.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 0.4 | GO:1902948 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.6 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 1.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 3.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.7 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 0.6 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.1 | 1.5 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.1 | 0.8 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.5 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.1 | 1.1 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 1.2 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 3.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.1 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 3.5 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.3 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 0.3 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 | 2.9 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 3.5 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.1 | 1.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 1.9 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 1.4 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 2.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 1.0 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.1 | 1.7 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.5 | GO:2000690 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.1 | 0.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 1.9 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 0.2 | GO:0006311 | meiotic gene conversion(GO:0006311) |
| 0.1 | 2.2 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 | 0.8 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 2.6 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 2.5 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 0.7 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 1.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.4 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 0.5 | GO:0001828 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.1 | 0.9 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.7 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 1.1 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.5 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 1.8 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.1 | 1.5 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 1.6 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 3.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 4.2 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.1 | 0.3 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.1 | 0.3 | GO:0090677 | response to mycophenolic acid(GO:0071505) cellular response to mycophenolic acid(GO:0071506) metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.1 | 1.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.3 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.8 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 1.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 0.7 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 2.8 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 0.6 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.1 | 1.4 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 0.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.5 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.3 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.1 | 0.5 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 1.4 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 0.1 | 0.9 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.7 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.7 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 1.7 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.1 | 1.1 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.1 | 0.1 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 2.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.8 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.1 | 0.2 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.1 | 0.5 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 4.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.5 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 1.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.5 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.9 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.1 | 0.6 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.4 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 1.7 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.4 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.2 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 2.1 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.1 | 4.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 2.4 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 3.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.5 | GO:1903052 | positive regulation of proteolysis involved in cellular protein catabolic process(GO:1903052) |
| 0.1 | 0.5 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 0.5 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.2 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.1 | 1.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 1.0 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 2.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.1 | 2.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.2 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.1 | 0.4 | GO:0035900 | isocitrate metabolic process(GO:0006102) response to isolation stress(GO:0035900) |
| 0.1 | 1.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.4 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.8 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.6 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.1 | GO:2000667 | positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.1 | 0.5 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 1.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.0 | 0.7 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.4 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.5 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.6 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.5 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 0.3 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.9 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.5 | GO:0060180 | female mating behavior(GO:0060180) |
| 0.0 | 0.6 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 3.8 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.5 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.3 | GO:0031620 | regulation of fever generation(GO:0031620) positive regulation of fever generation(GO:0031622) |
| 0.0 | 1.2 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.3 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.6 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 1.2 | GO:0045187 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.4 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 1.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.8 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 1.8 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 1.1 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 4.7 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.6 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.3 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 1.4 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.6 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 2.3 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.7 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.2 | GO:0098704 | fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.6 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 1.3 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.0 | 0.3 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 1.3 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 4.1 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.6 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.1 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.0 | 0.3 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.3 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 3.1 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 0.2 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.8 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 2.5 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.2 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 3.5 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.5 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.9 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 1.0 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 1.7 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.4 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 0.7 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.7 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.0 | 0.8 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 1.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.2 | GO:0045329 | carnitine metabolic process(GO:0009437) carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.9 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.4 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.8 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.6 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 2.2 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.1 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.1 | GO:0006612 | protein targeting to membrane(GO:0006612) cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.3 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.0 | 0.1 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.0 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 1.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.7 | GO:1901031 | regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.0 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.0 | 0.2 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.4 | GO:0061458 | reproductive system development(GO:0061458) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.4 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.6 | GO:0001501 | skeletal system development(GO:0001501) |
| 0.0 | 0.1 | GO:0061744 | psychomotor behavior(GO:0036343) motor behavior(GO:0061744) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0003170 | heart valve development(GO:0003170) |
| 0.0 | 0.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 3.0 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.0 | GO:0006043 | glucosamine catabolic process(GO:0006043) |
| 0.0 | 0.3 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.0 | 1.2 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 1.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.2 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.8 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 1.2 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 1.8 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.0 | GO:0009750 | response to fructose(GO:0009750) |
| 0.0 | 0.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.7 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 1.3 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.6 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.9 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.9 | 10.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.6 | 4.5 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.5 | 4.8 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.5 | 4.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.5 | 1.5 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.4 | 2.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.4 | 5.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.4 | 1.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.4 | 6.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.3 | 2.0 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.3 | 1.0 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.3 | 9.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 1.9 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.3 | 3.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.3 | 1.8 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.3 | 0.9 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.3 | 1.4 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.3 | 1.4 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.3 | 1.5 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.2 | 18.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 2.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 3.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 1.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 6.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 0.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.2 | 1.8 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.2 | 0.6 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 3.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 1.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.2 | 1.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 5.4 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.2 | 0.6 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.9 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.4 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 2.1 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.1 | 2.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.8 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.5 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.1 | 1.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.9 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.5 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 4.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 2.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.4 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 2.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.6 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.4 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 2.5 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 0.6 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.1 | 1.0 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 1.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.8 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 1.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.3 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.1 | 2.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 13.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 1.3 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 6.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 7.3 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.1 | 1.4 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 1.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 1.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.8 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.7 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 1.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 2.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.2 | GO:0070877 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.1 | 2.2 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 4.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.9 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 0.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 3.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.0 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.4 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 1.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.6 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 1.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.6 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.7 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 1.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.9 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.5 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 3.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 6.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 1.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 2.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.9 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 1.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.1 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.1 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 2.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 1.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 1.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.4 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 1.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 1.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 4.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 2.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 5.0 | GO:0045121 | membrane raft(GO:0045121) |
| 0.0 | 1.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 2.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 1.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 2.0 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.5 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0002102 | podosome(GO:0002102) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.4 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.7 | 5.5 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.7 | 2.6 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.6 | 5.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.6 | 1.8 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.6 | 4.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.5 | 4.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.4 | 2.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.4 | 2.4 | GO:0052848 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.4 | 2.4 | GO:0017153 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.4 | 1.6 | GO:0090556 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.4 | 4.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.4 | 1.1 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.4 | 5.3 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.3 | 1.0 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.3 | 4.4 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.3 | 1.7 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.3 | 1.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.3 | 4.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.3 | 1.6 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.3 | 1.9 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.3 | 2.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.3 | 1.7 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.3 | 5.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.3 | 2.6 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.3 | 1.2 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.3 | 1.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.3 | 1.3 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.3 | 6.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.3 | 2.5 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.2 | 3.5 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 1.0 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.2 | 0.9 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.2 | 1.4 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.2 | 0.9 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 3.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 1.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.2 | 2.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.2 | 5.7 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.2 | 0.8 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 11.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 0.9 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.2 | 2.4 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 1.6 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.2 | 1.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.2 | 2.7 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.2 | 0.5 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.2 | 1.5 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.2 | 2.2 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 1.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.2 | 0.6 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.2 | 0.8 | GO:0089720 | caspase binding(GO:0089720) |
| 0.2 | 2.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 2.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.4 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 1.4 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 1.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.4 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 1.7 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.1 | 4.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 3.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 4.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.5 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.2 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 1.9 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 1.0 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.2 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.1 | 0.7 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 1.7 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 4.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.3 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.1 | 5.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.9 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.2 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.1 | 1.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.5 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.1 | 5.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.7 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.4 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.1 | 0.7 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.3 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 1.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 1.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 5.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.6 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 1.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.7 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.9 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.5 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.5 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 1.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 1.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 1.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 2.5 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.1 | 17.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.6 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.4 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 0.6 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.3 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.1 | 0.6 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.9 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 1.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.1 | 0.4 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.5 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 0.2 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 0.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.2 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 1.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 1.6 | GO:0055103 | ligase regulator activity(GO:0055103) |
| 0.1 | 1.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 1.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 2.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.6 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.6 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 2.1 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 3.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.2 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 2.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.6 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 0.2 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.1 | 0.1 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.1 | 2.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.6 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 7.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 13.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 1.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 1.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 1.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 3.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.3 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 2.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 1.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 1.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 5.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.9 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 1.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.4 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.9 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 2.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.8 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 1.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 1.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.9 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 7.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 5.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.0 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.2 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 1.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 1.0 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.4 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 3.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 1.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.6 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 3.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.8 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.5 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 5.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.9 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.0 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.3 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.0 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.0 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 2.0 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 10.8 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.0 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 4.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.6 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.3 | GO:0031369 | translation initiation factor binding(GO:0031369) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 10.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 16.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 6.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 4.5 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 3.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 3.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 5.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 10.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 9.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 8.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 3.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 6.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.0 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.8 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 5.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 6.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 4.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.8 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 1.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 13.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 4.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 3.3 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 2.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 3.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.9 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.4 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.4 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.3 | 3.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.3 | 6.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 4.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 29.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.2 | 5.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 5.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 4.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 5.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 9.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 2.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 4.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 1.7 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 1.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 2.0 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 3.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.0 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 2.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 9.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 5.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 3.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 4.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 4.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 1.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 0.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.1 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 1.9 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 1.1 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 0.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 1.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.0 | 2.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.9 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.0 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 5.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.6 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 3.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.9 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.0 | 2.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 2.4 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 1.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.3 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 1.7 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 1.1 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 1.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 4.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 4.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |